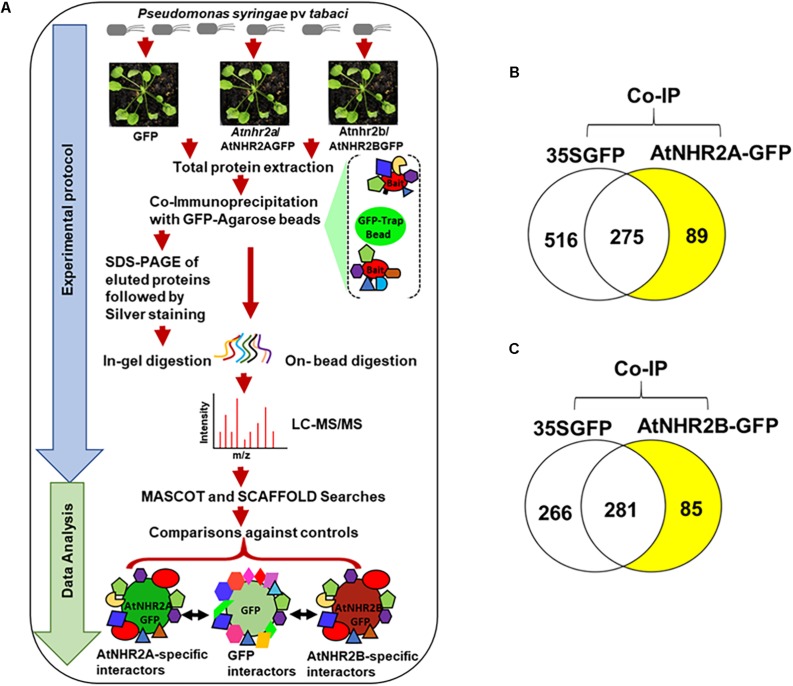
FIGURE 1.
Co-immunoprecipitation/mass spectrometry analysis to unravel the AtNHR2A and AtNHR2B Interactomes. Co-IP/MS workflow summarizing experimental protocol and data analysis. For the experimental protocol, four-week-old transgenic Arabidopsis thaliana plants expressing AtNHR2A-GFP and AtNHR2B-GFP GFP were inoculated with Pseudomonas syringae pv tabaci at 1 × 106 CFU/mL. At 6 hpi, plants were harvested for total protein extraction. Extracted proteins from each sample were subjected to co-immunoprecipitation using GFP Trap A beads. GFP-bound protein complexes were eluted with 2X SDS sample buffer and resolved by SDS-PAGE for silver staining and in-gel trypsin digestion, or directly processed for on-bead trypsin digestion. Both in-gel and on-bead tryptic digested products were analyzed by LC-MS/MS. For data analysis, identified peptide spectra were then analyzed by Mascot using the Arabidopsis thaliana protein database as reference, and identified peptides were further validated and compared using Scaffold to identify peptides representing proteins that specifically interact with AtNHR2A-GFP or AtNHR2B-GFP but not with GFP. Peptide identification were done by setting peptide hits ≥ 2 with the false discovery rate (FDR) below 1% based on decoy database (A). Venn diagrams show specific proteins interacting with AtNHR2A-GFP (B) and AtNHR2B-GFP (C) (shaded in yellow).