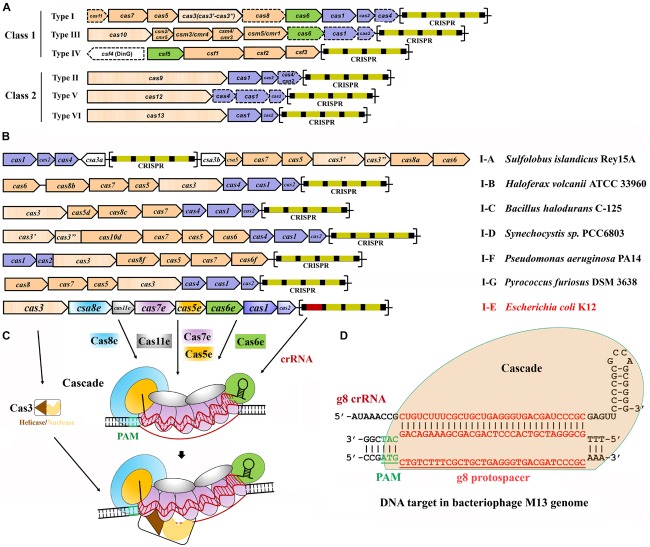
FIGURE 1.
Classification and architecture of CRISPR-Cas systems and interference by type I systems. (A) CRISPR-Cas systems are greatly diverse and can be classified into two classes, class 1 and class 2. Class 1 systems encode multisubunit effector complexes; class 2 systems encode single-subunit effectors. Genes that may miss in certain subtypes are indicated with dashed outlines. Genes encoding the components of each interference complex are colored in orange, and those involved in crRNA processing and new spacer acquisition are in green and blue, respectively. The effector nucleases for each subtype are shown with filled vertical lines. CRISPR arrays are indicated, with squares and rectangles representing repeats and spacers, respectively. (B) Organization of the CRISPR-Cas loci for the typical type I subtypes. Representative operons for each type are shown, and gene names are indicated. Gene functions are marked with colors the same as shown in (A), except for the subtype I-E of Escherichia coli K12. CRISPR arrays are indicated, with squares and rectangles representing repeats and spacers, respectively. (C) Schematic of DNA targeting by the representative type I-E of E. coli K12. It is composed of a crRNA bound by Cas5 and Cas6 at either end and Cas7 subunits along the guide region, a large subunit (Cas8), and sometimes a small subunit (Cas11). Upon PAM recognition by Cas8, Cascade binding to the target DNA leads to DNA duplex destabilization, allowing crRNA invasion to form a full R-loop. Cas3 is recruited to the R-loop and nicks the replaced strand of the target DNA within the protospacer. (D) Schematic showing type I-E Cascade containing a crRNA (g8 crRNA) targeting a sequence on the genome of bacteriophage M13. Sequences of PAM and protospacer are indicated with underlined green and red fonts, respectively [constructed according to Semenova et al. (2011)].