FIG 10.
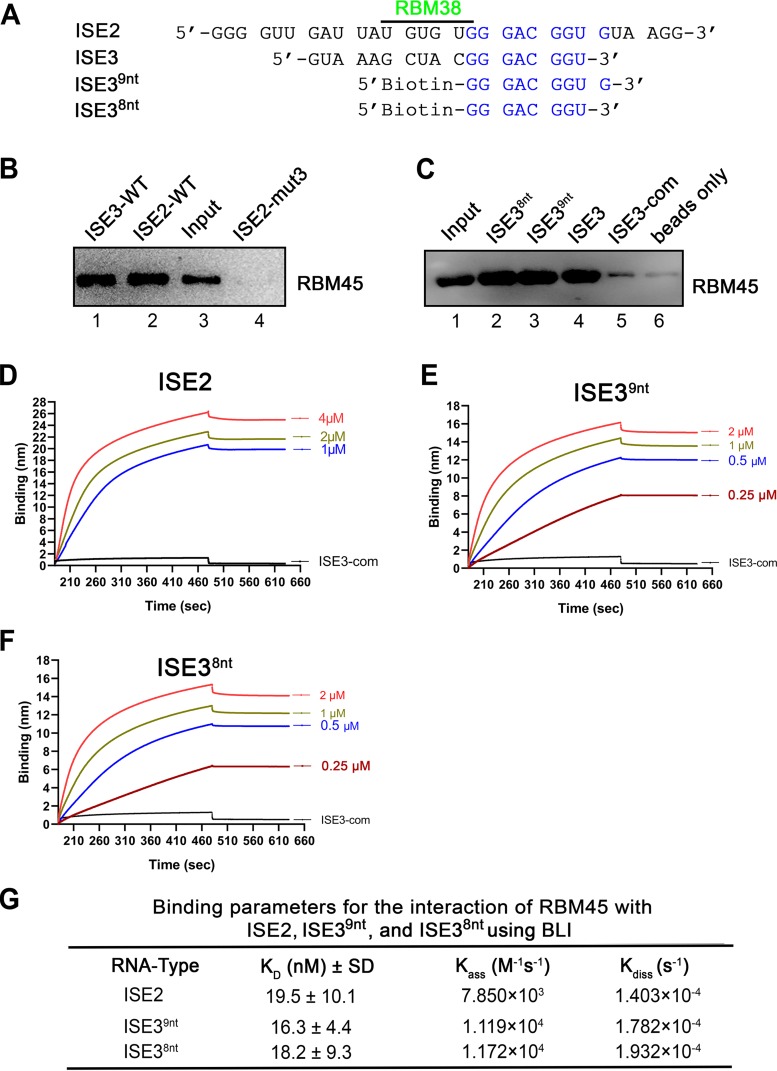
ISE2 specifically binds the RBM45 protein under in vitro conditions. (A) Alignment of the ISE2 and ISE3 sequences. ISE2 and ISE3 share the 8 nucleotides 5′-GGG ACG GU-3′ indicated in blue. (B and C) RNA pulldown assay. Biotinylated ISE2 and ISE3 and ISE2-mut3 (negative control) (B) and biotinylated ISE38nt, ISE39nt, ISE3, and ISE3-com (C) were incubated with recombinant RBM45-His protein and then pulled down by using streptavidin-conjugated beads. Eluted RNA-bound proteins were analyzed by Western blotting for RBM45. A 3-μg volume of the recombinant RBM45-His protein was loaded as the input. (D to F) Biolayer interferometry (BLI). Data represent results of comparisons of binding affinities of the RBM45 protein at the indicated concentration for ISE2 RNA (D), ISE39nt (E), and ISE38nt (F). BLI sensorgrams show association and dissociation of the RBM45 protein with ISE2 RNA (D), ISE39nt (E), and ISE38nt (F) at different concentrations over time as indicated. (G) KD determination. Binding parameters used to calculate KD values and ratios of dissociation (Kass) and association (Kdiss) rate constants. The binding experiments were repeated at least three times for calculating the means and standard deviations (SD), by using various concentrations of the protein.