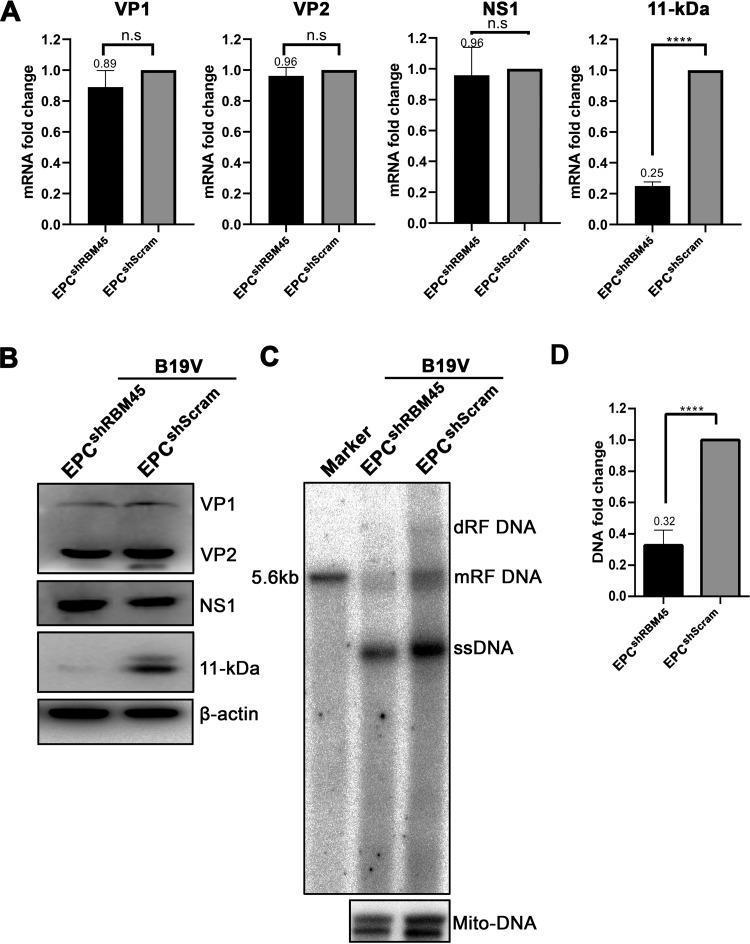
FIG 3.
RBM45 regulated 11-kDa protein expression in CD36+ EPCs. CD36+ EPCs transduced with shRBM45-expressing and scramble shRNA (shScram)-expressing lentiviruses were infected with B19V at 2 days postransduction. The cells were collected at 2 days postinfection for analyses. (A) RT-qPCR. Total RNA was extracted. cDNAs reverse transcribed from the RNAs were used for qPCR of each viral mRNA, as indicated, using specific primers and probes for each viral mRNA as described previously (37). The quantified viral mRNA level was normalized to the level of β-actin mRNA. The values representing viral mRNA from B19V-infected CD36+ EPCs transduced with shScram were used as controls and arbitrarily set to 1. (B) Western blotting. CD36+ EPCs were collected at 2 days postinfection, lysed, and run for Western blotting. Blots were probed for the VP1, VP2, NS1, and 11-kDa proteins, using their respective antibodies. Blots were reprobed for β-actin as a loading control. (C and D) Southern blotting. At 2 days postinfection, Hirt DNA was extracted from CD36+ EPCs for Southern blot analysis. (C) The blots were probed with the M20 probe (top) and the mitochondrial DNA probe (Mito-DNA) (bottom), respectively. Representative blots are shown. dRF, mRF, and ssDNA, double replicative form, monomer replicative form, and single-stranded DNA, respectively. A 10-ng volume of SalI-digested M20 was used as a size marker of 5.6 kb. (D) The intensity of the RF DNA band was quantified and normalized to the level of the mitochondrial DNA (Mito-DNA) of each sample. The value representing viral RF DNA in B19V-infected CD36+ EPCs transduced with shScram was arbitrarily set to 1. Values representing relative fold change of the viral RF DNA in B19V-infected CD36+ EPCs transduced with shRBM45 are shown with averages and standard deviations from three repeated experiments.