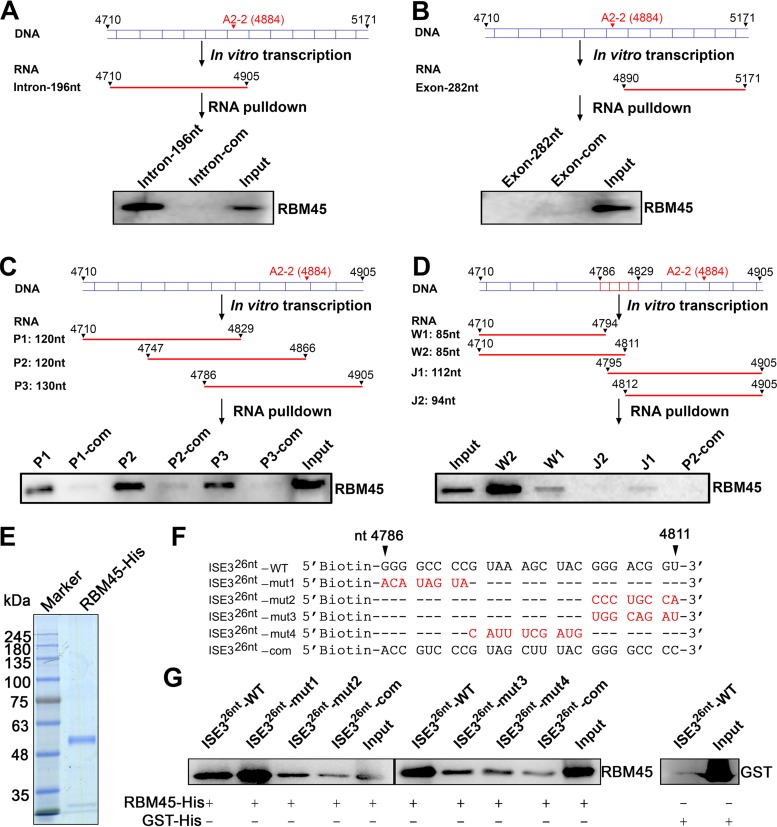
FIG 4.
Identification of a novel intronic splicing enhancer in B19V pre-mRNA that was bound by RBM45 specifically. (A to D) Schematic representations of in vitro transcription and RNA pulldown assay. The B19V DNA region surrounding the A2-2 site is shown. Biotin-labeled in vitro-transcribed RNA (shown in red) was captured by streptavidin-conjugated agarose beads, followed by incubation with UT7/Epo-S1 nuclear lysates. RNA-bound proteins were eluted and analyzed by Western blotting. (A) B19V DNA (nt 4710 to 4905) was used as a template to transcribe RNA (named 11-kDa intron), while its cRNA (named intron com) was transcribed as a negative control. (B) B19V DNA (nt 4890 to 5171) was used as a template to transcribe RNA (named 11-kDa exon), while its cRNA (named exon com) was transcribed as a negative control. (C) B19V DNA (nt 4710 to 4905) was in vitro transcribed to generate 3 overlapped small RNA segments termed RNA P1 (nt 4710 to 4829), P2 (nt 4747 to 4866), and P3 (nt 4786 to 4905). cRNA P1, P2, and P3 (complementary P1 [P1-com], P2-com, and P3-com, respectively) were transcribed as negative controls. (D) B19V DNA (nt 4710 to 4905) was further in vitro transcribed to generate 4 small segments termed RNA W1 (nt 4710 to 4794), W2 (nt 4710 to 4811), J1 (nt 4795 to 4905), and J2 (nt 4812 to 4905), while P2 cRNA (P2-com) was transcribed as a negative control. (E to G) RNA pulldown assay using biotinylated ISE326nt WT and its mutants. Recombinant RBM45-His protein (E) was incubated with the corresponding biotinylated RNA oligonucleotides (F), pulled down with streptavidin-conjugated beads, and analyzed by Western blotting for RBM45 (G). For panel E, A 6-μg volume of the RBM45-His protein was run on an SDS-10% PAGE gel and then stained with Coomassie brilliant blue dye. Complementary ISE326nt (ISE326nt-com) and GST-His served as the negative RNA and protein controls, respectively.