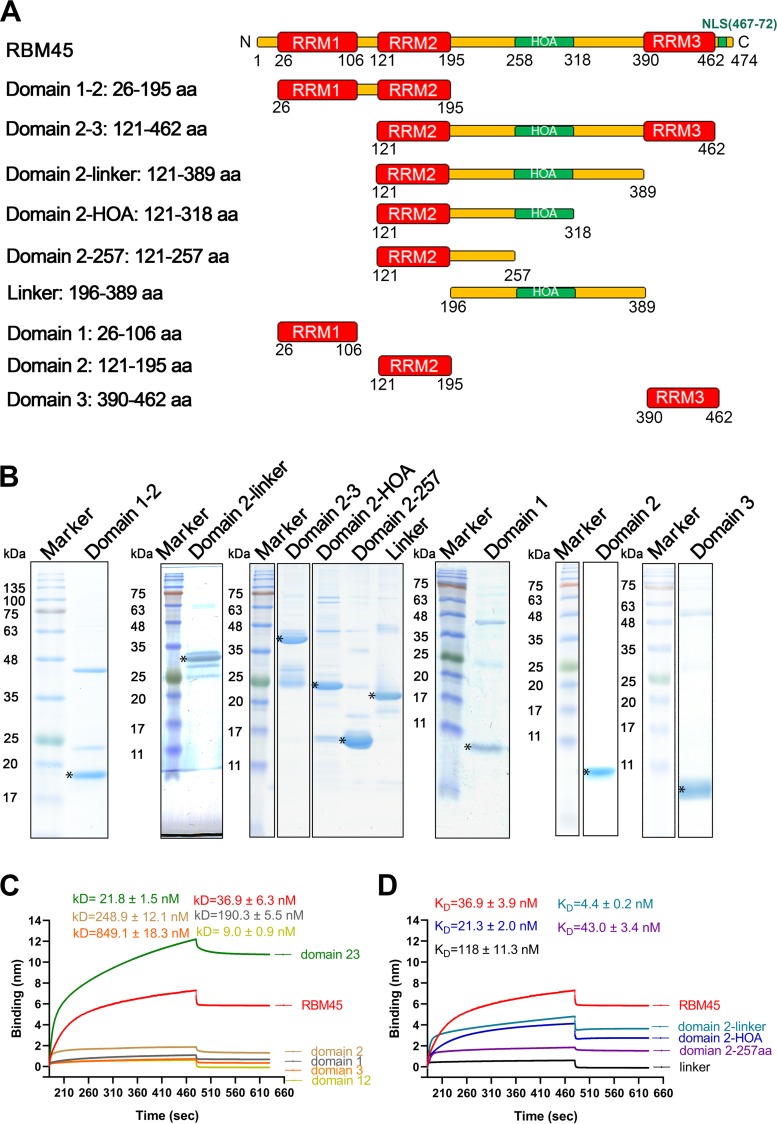
FIG 6.
The RRM2-HOA domain of RBM45 was largely responsible for binding to ISE3 of B19V pre-mRNA. (A) Domain structures of RBM45. The RNA recognition motif (RRM) domains are shown in red. The homo-oligomer assembly (HOA) domain and nuclear localization sequence (NLS; residues 469 to 472) are shown in green. Linker regions between RRMs are diagrammed in yellow. (B) Purification of truncated RBM45 proteins. Purified recombinant proteins (6 μg) were separated on an SDS-10% or 15% PAGE gel and stained with Coomassie brilliant blue dye. Asterisks denote the truncated proteins. (C and D) Biolayer interferometry (BLI) analysis. We used BLI to measure the interactions of ISE3 with different truncated and full-length forms of RBM45. (C) Domain 1–2 (aa 26 to 195), domain 2–3 (aa 121 to 462), domain 1 (aa 26 to 106), domain 2 (aa 121 to 195), and domain 3 (aa 390 to 462) of RBM45 were analyzed for their binding capability with ISE3. Domain 2–3 (aa 121 to 462) was found to be the key domain for RBM45 binding to ISE3. (D) Data representing domain 2-linker (aa 121 to 389) and 2-HOA (aa 121 to 318) were analyzed and compared with data representing binding to the full-length RBM45. KD values are shown for each binding curve. The binding experiments were repeated at least three times for calculating means and standard deviations (SD).