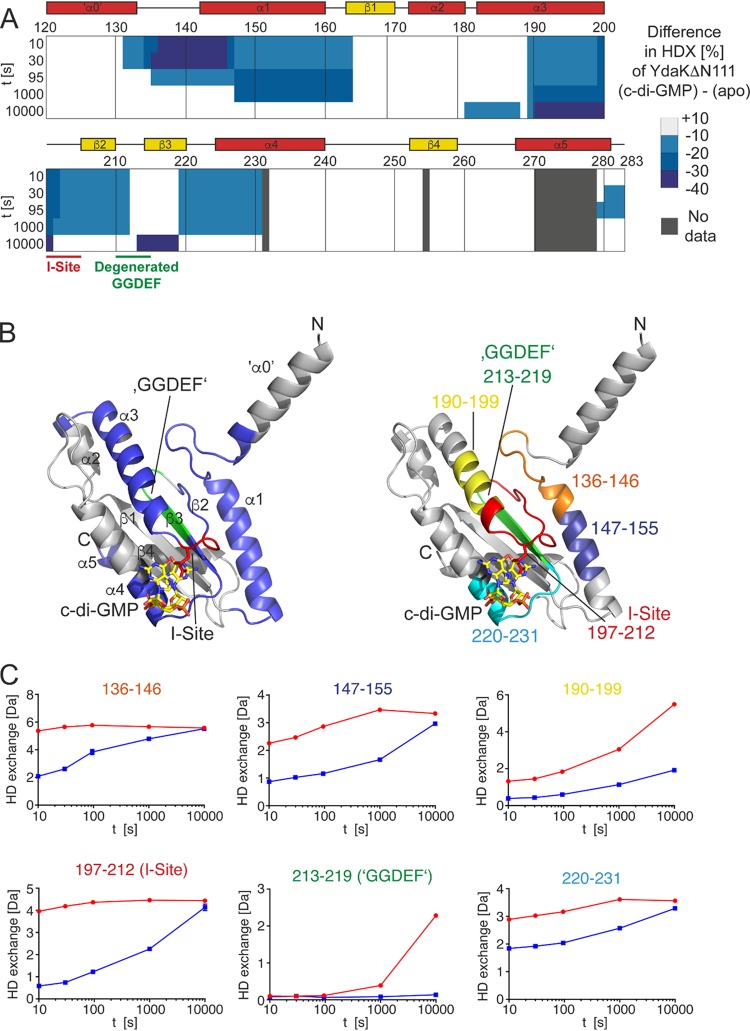
FIG 5.
c-di-GMP alters the conformation of YdaK. (A) Amino acid residues of YdaK-ΔN111 are colored according to their differences in HDX profiles between c-di-GMP-bound YdaK-ΔN111 and apo-YdaK-ΔN111. The secondary structure of YdaK-ΔN111 based on a generated model is indicated. (B, left) Locations of regions with less HDX in the presence of c-di-GMP in a structural model of YdaK-ΔN111. The I-site and degenerated GGDEF motifs are in red and green, respectively. The I-site arginine 202 is shown as sticks. The position of c-di-GMP bound to the I-site is inferred from a superimposition of the YdaK-ΔN111 structural model upon the crystal structure of the GGDEF domain of Dcsbis from Pseudomonas aeruginosa (PDB accession number 4ZMM). (Right) Location of representative peptides in the structural model of YdaK-ΔN111. (C) Hydrogen/deuterium exchange profiles of representative YdaK peptides in the c-di-GMP-bound (red) and unliganded (blue) states. Data represent the means ± standard deviations (SD) of results from three technical replicates. t, time.