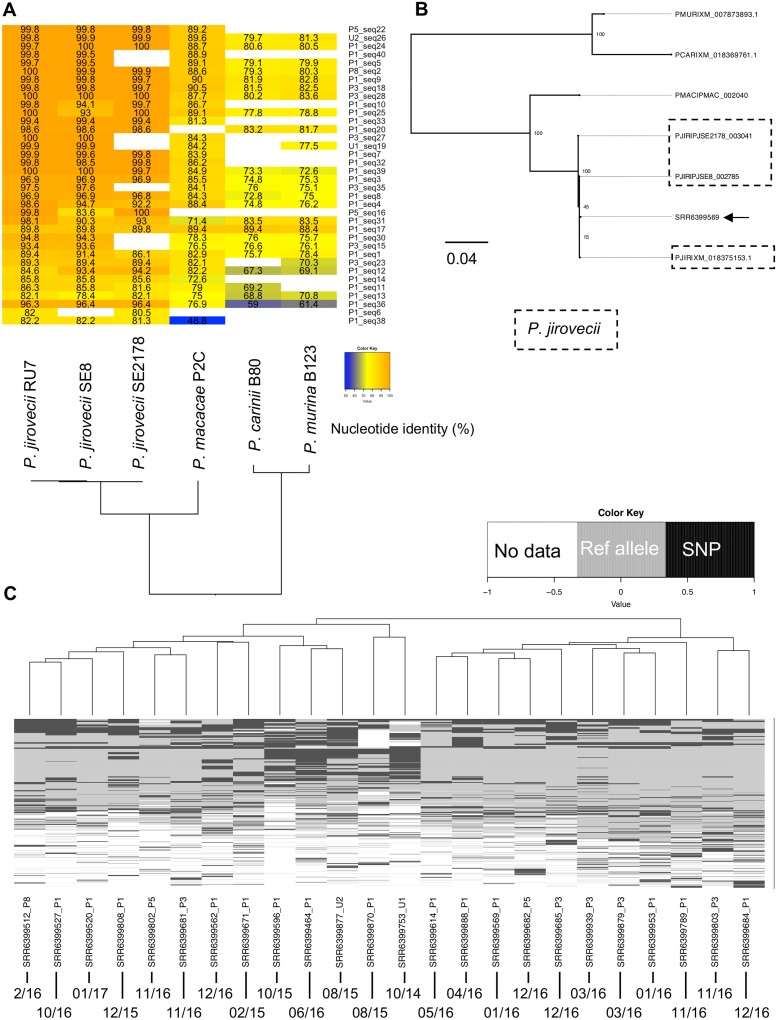
FIG 1.
Evidence of Pneumocystis DNA in air metagenomes. (A) Pairwise nucleotide identity heatmap of 37 of 45 contigs. Only contigs for which a 1-to-1 ortholog could be established were used. Blank spaces in the heatmap represent a lack of an identifiable ortholog. These missing orthologs likely represent an incomplete genome or annotation rather than real biological gene loss. (B) Maximum likelihood phylogeny of a representative contig encoding the 3-oxoacyl-[acyl-carrier-protein] synthase (arrow). The contig was recovered from the NCBI SRA accession number SRR6399569. Bootstrap values of 100 replicates are presented on the tree nodes. Sequences are labeled with the following species identifiers: PJIR, Pneumocystis jirovecii; PMAC, Pneumocystis sp. macacae; PCAR, P. carinii; PMUR, P. murina. P. jirovecii sequences are boxed. (C) Single nucleotide polymorphisms (SNP) were identified from the alignments of raw reads from each SRA file to P. jirovecii reference genome assembly strain RU7. The heatmap shows the variant calling results for 1,852 genomic positions encoded as follows: black, presence of a SNP; gray, region covered by at least two sequencing reads with the allele as the reference genome; white, absence of coverage. Each row represents a unique SNP, and each column represents the 15 samples with detectable Pneumocystis sequences. Collection dates (month/year) are presented at the bottom of the figure.