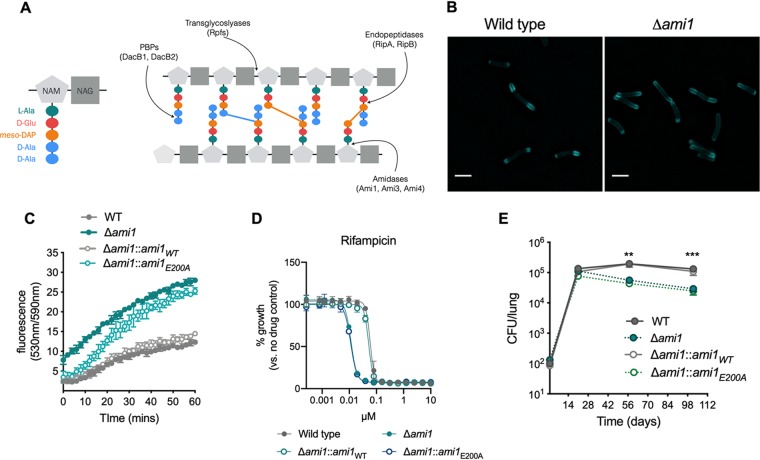
FIG 1.
The peptidoglycan amidase Ami1 is dispensable for M. tuberculosis cell division in vitro but is important for persistence in vivo. (A) Schematic diagram of peptidoglycan (PG). N-Acetyl muramic acid (NAM) and N-acetylglucosamine (NAG) residues make up the glycan backbone of PG. The pentapeptide stem constitutes of l-alanine, d-glutamate, meso-diaminopimelic acid (mDAP), and two d-alanine residues (left panel). Different classes of PG-hydrolyzing enzymes cleave the PG macromolecule at different sites (right panel). The colored bars between peptide stems represent cross-links (blue, 4-3; orange, 3-3). (B) Micrographs of M. tuberculosis cells labeled with the fluorescent d-alanine analogue HADA for 20 h. Bar, 2 μm. (C) Ethidium bromide (EtBr) uptake assay. Bacteria were incubated with EtBr, and the fluorescence (indicating uptake of EtBr and binding to DNA) was monitored over the course of 60 min. (D) MIC profiles of Δami1, Δami1::ami1WT, and Δami1::ami1E200A strains relative to that of the wild type for the frontline TB drug rifampin. (E) Bacterial burden in the lungs of CB57BL/6 mice infected by aerosol inhalation with the following strains: wild-type H37Rv (WT), Δami1, Δami1::ami1WT, and Δami1::ami1E200A variant. Significance was determined by one-way analysis of variance (ANOVA) and adjusted for multiple comparisons. **, adjusted P (adj-P) < 0.005; ***, adj-P < 0.0005. All data are representative of two independent experiments. Data in panel E are from 2 independent experiments, each with 4 mice per group and time point.