Figure 6.
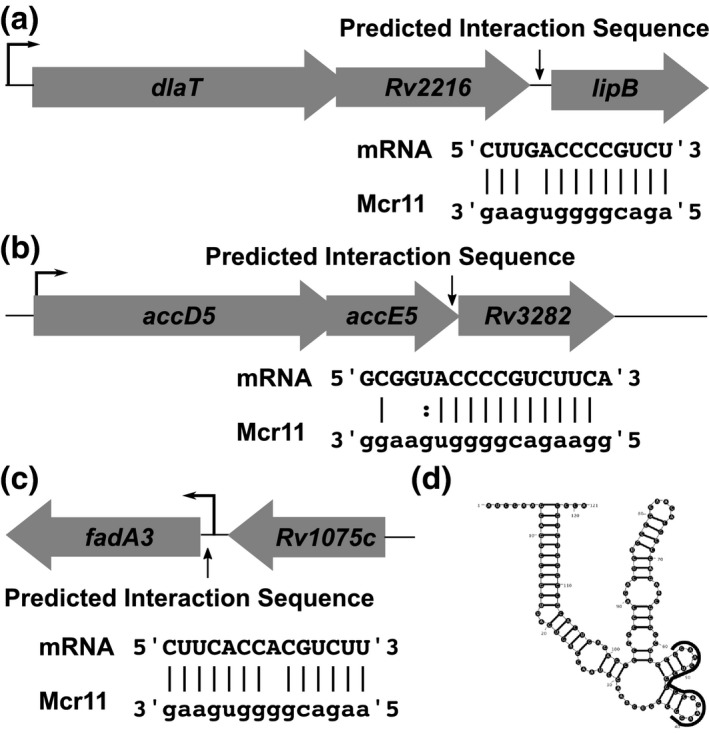
Bioinformatic modeling of Mcr11 targets reveals potential regulatory targets that are involved in central metabolism and cell division. (a) The organization of the dlaT‐lipB locus, with the position and potential base‐pairing interactions between the mRNA and Mcr11 is shown below. The transcriptional start site of the operon is shown with a thin black arrow. (b) As in (a), but for the accD5‐Rv3282 locus. Dashes indicate Watson–Crick base pairs, dots indicate non‐Watson–Crick base pairs, and a blank space indicates no interaction between bases. (c) As in (a) and (b), but for the Rv1074c/fadA3 locus. (d) The MFE secondary structure of Mcr11 as predicted by RNAStructure, with the portion of the sRNA predicted to interact with targets is shown in (a‐c) outlined in black
