Abstract
In this study, we examined the Ca2+‐permeable Piezo1 channel, a newly identified mechanosensing ion channel, in human dental pulp‐derived mesenchymal stem cells (MSCs) and hypothesized that activation of the Piezo1 channel regulates MSC migration via inducing ATP release and activation of the P2 receptor purinergic signaling. The Piezo1 mRNA and protein were readily detected in hDP‐MSCs from multiple donors and, consistently, brief exposure to Yoda1, the Piezo1 channel‐specific activator, elevated intracellular Ca2+ concentration. Yoda1‐induced Ca2+ response was inhibited by ruthenium red or GsMTx4, two Piezo1 channel inhibitors, and also by Piezo1‐specific siRNA. Brief exposure to Yoda1 also induced ATP release. Persistent exposure to Yoda1 stimulated MSC migration, which was suppressed by Piezo1‐specific siRNA, and also prevented by apyrase, an ATP scavenger, or PPADS, a P2 generic antagonist. Furthermore, stimulation of MSC migration induced by Yoda1 as well as ATP was suppressed by PF431396, a PYK2 kinase inhibitor, or U0126, an inhibitor of the mitogen‐activated protein kinase MEK/ERK signaling pathway. Collectively, these results suggest that activation of the Piezo1 channel stimulates MSC migration via inducing ATP release and subsequent activation of the P2 receptor purinergic signaling and downstream PYK2 and MEK/ERK signaling pathways, thus revealing novel insights into the molecular and signaling mechanisms regulating MSC migration. Such findings provide useful information for evolving a full understanding of MSC migration and homing and developing strategies to improve MSC‐based translational applications.
Keywords: ATP release, MEK/ERK, P2 receptor, Piezo1 channel, PYK2
A, Exposure to Yoda1, the Piezo1 channel‐specific activator, stimulates human dental pulp mesenchymal stem cell migration in wound healing assay; B, the underlying signaling mechanisms.

Significance statement.
Mesenchymal stem cells (MSCs) release ATP as an extracellular signaling molecule in response to mechanical and diverse other stimuli, but the underlying mechanisms remain largely elusive. This study shows the expression of the Ca2+‐permeable Piezo1 channel, a newly discovered mechanosensing mechanism, in human dental pulp‐derived MSCs and its important role in the regulation of MSC migration. It further demonstrates that activation of the Piezo1 channel stimulates MSC migration via inducing ATP release and subsequent activation of P2 receptor purinergic signaling. This study thus provides novel insights into the mechanisms regulating MSC migration. Such information is useful for gaining a better understanding of MSC migration and designing strategies to improve applications of MSCs in tissue engineering and regenerative medicine.
1. INTRODUCTION
Mesenchymal stem cells (MSCs), which have been isolated from bone marrow, umbilical cord, dental pulp, and a diversity of other tissues, have the ability of differentiating into tissue‐specific lineages.1, 2, 3 An increasing number of studies have demonstrated promising applications of MSCs in regenerative medicine and tissue engineering to repair and regenerate damaged or lost tissues due to diseases and traumatic injuries4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21 and, nonetheless, translational uses of MSCs remain inefficacious, in part due to their limited capability of migrating and homing.3, 22, 23, 24, 25, 26 Thus, gaining insights into the mechanisms regulating MSC migration can provide useful information for developing novel strategies to prepare MSCs for clinical applications and improve the efficacy of MSC‐based therapies as well as evolving a better understanding of the contentious MSC biology.27, 28 A multiplicity of extracellular signaling molecules and cell surface receptors for them and coupled intrinsic signaling mechanisms have been shown to regulate cell migration.29, 30, 31, 32 ATP represents one such signaling molecule that selectively activates the P2 purinergic receptor family, which is composed of ligand‐gated ion channel P2X receptors and G‐protein‐coupled P2Y receptors.32, 33 ATP‐induced purinergic signaling has been well documented in MSCs from different species and tissues34 and, furthermore, has been demonstrated to play an important role in regulating MSC proliferation35, 36 and differentiation37, 38 as well as migration and homing.39, 40
Increasing evidence indicates that various physical and chemical stimuli can stimulate ATP release from MSCs.35, 38 MSCs are mechanosensitive,41, 42 and mechanical forces can induce ATP release, but the mechanisms for mechanical induction of ATP release from MSCs remain poorly understood.32 The Ca2+‐permeable Piezo1 channel is a newly discovered mechanosensing mechanism in many types of cells.43, 44, 45, 46, 47, 48, 49, 50, 51, 52, 53, 54, 55 Furthermore, recent studies have reported that mechanical or chemical activation of the Piezo1 channel induces ATP release from endothelial cells,48 urothelial cells,54 and red blood cells.55 There is also compelling evidence to support a critical role for the Piezo1 channel in regulating cell migration in endothelial cells,56 breast cancer cells,57 gastric cancer cells,50, 58 and small lung cancer cells.59 Recent studies have also shown the expression of the Piezo1 channel in human bone marrow‐derived MSCs (BM‐MSCs)60 and rat dental pulp‐derived stem cells (DP‐MSCs).61 Therefore, it is attractive to hypothesize that the Piezo1 channel regulates MSC migration via inducing ATP release and subsequent activation of the P2 receptor purinergic signaling.32, 40
Recent studies have reported that the mitogen‐activated protein kinase/extracellular signal‐regulated kinase (MEK/ERK) or ERK is critically engaged as a downstream signaling pathway in mediating acetylcholine‐induced rat BM‐MSC migration62 and interleukin‐1β‐induced human umbilical cord‐derived MSC (UC‐MSC) migration.63 It has been proposed that the Piezo1 channel mediates ultrasound‐induced activation of the MEK/ERK signaling pathway in rat DP‐MSCs.61 The proline‐rich tyrosine kinase 2 (PYK2) is well‐known as a signaling molecule downstream of intracellular Ca2+ and plays a critical role in mediating Ca2+‐dependent induction of the MEK/ERK pathway in monocyte, microglial, and neuronal cells.64, 65, 66, 67, 68, 69, 70 There is evidence for involvement of PYK2 in cancer cell migration,70 but it is unclear whether PYK2 plays a role in the regulation of MSC migration.
Therefore, in this study, we examined the expression of the Piezo1 channel in human dental pulp‐derived MSCs (hDP‐MSCs) and its role in the regulation of cell migration. Particularly, we investigated the hypothesis of activation of the Piezo1 channel as a mechanism regulating hDP‐MSC migration via inducing ATP release and subsequent activation of the P2 receptor purinergic signaling. We also explored the role of PYK2 and MEK/ERK as the downstream signaling pathways in mediating hDP‐MSC migration. Our results show functional expression of the Piezo1 channel in hDP‐MSCs and its important role in stimulating cell migration, and provide further evidence to support that activation of the Piezo1 channel stimulates cell migration via inducing ATP release and subsequent activation of the P2 receptor purinergic signaling and downstream PYK2 and MEK/ERK signaling pathways. These novel findings provide a better understanding of the molecular and signaling mechanisms regulating MSC migration and useful information for the development of strategies to improve applications of MSCs in regenerative medicine and tissue engineering.
2. MATERIALS AND METHODS
2.1. Chemicals and media
All chemicals or reagents were obtained from Sigma‐Aldrich, unless specified otherwise. Yoda1, PF431396, GsMTx4, and PPADS were from Tocris. U0126 and ionomycin were from Cayman Chemical. Ruthenium red (RR) was from Merck. Ca2+/Mg2+‐free phosphate‐buffered saline (PBS), Dulbecco's modified Eagle's medium (DMEM), GlutaMAX‐I, OPTI‐MEM, fetal bovine serum (FBS), penicillin‐streptomycin, trypsin‐EDTA, Fura‐2/acetoxymethyl (Fura‐2/AM), and pluronic acid F‐127 were from Invitrogen.
2.2. Cell culture
Human DP‐MSCs were isolated from the molar teeth from two female donors of 9 and 32 years old and two male donors of 22 and 20 years old (9F, 32F, 22M, and 20M in abbreviation) and characterized for the expression of positive and negative cell surface MSC markers and in vitro differentiation potentials as described in our previous study.40 Cells were maintained in DMEM supplemented with GlutaMAX‐I, 10% FBS and 100 units/mL penicillin and 100 μg/mL streptomycin at 37°C and 5% CO2 in a tissue culture incubator. Cells, when reaching 80‐90% confluency, were passaged and, when cells were settled down during incubation for 24 hours, the growth medium was replaced with fresh growth medium. Cells of 4‐6 passages were used in this study.
2.3. RT‐qPCR analysis
Cells were plated at 80 000 per well in six‐well plates. Cells were collected when reaching 80‐90% confluency, and total RNA was extracted using a RNAqueous‐Micro kit and treated with a TURBO DNA‐free kit (both from Invitrogen). The RNA concentration and purity were determined using a Nanodrop 2000c spectrophotometer (Thermo Scientific), and 0.6 μg RNA was reverse‐transcribed into cDNA in a 20‐μL reaction volume using a High Capacity RNA‐to‐cDNA kit (Applied Biosystems) and a Mastercycler Gradient PCR machine (Eppendorf) at 37°C for 60 minutes. The reverse transcription (RT) was stopped at 95°C for 5 minutes. The resulting cDNA was stored at −20°C for real‐time polymerase chain reaction (qPCR) using a SensiMix SYBR & Fluorescein kit (Bioline) according to the manufacturer's instructions. A total of 20 μL of qPCR sample contained 50 ng/μL cDNA, SenisMix SYBR & Fluorescein, 0.25 μM forward primer, and 0.25 μM reversal primer. The primer sequences used are 5′‐AGATCTCGCACTCCAT‐3′ (forward) and 5′‐CTCCTTCTCACGAGTCC‐3′ (reverse) for Piezo1, and 5′‐TTGAGACCTTCAACACCC‐3′ (forward) and 5′‐TCTCTTGCTCGAAGTCC‐3′ (reverse) for β‐actin. Real‐time PCR was performed using a ROTOR‐Gene model 6000 machine (Corbett Research), consisting of 95°C for 10 minutes, 45 cycles of 95°C for 10 seconds, 60°C for 15 seconds, and 72°C for 20 seconds, followed by melting test from 72°C to 95°C. Data were analyzed using the ROTOR‐Gene 6000 series software. The PCR products with the anticipated size were confirmed by melting curve analysis. The cycle threshold value (Ct), representing the minimal cycle number at which the fluorescence signal grows above the background level,71 was set at 0.2. The mRNA expression level of Piezo1 was normalized to that of β‐actin based on the following equation72: 2[−(Ct Piezo1 −Ct β‐actin )]. The PCR products were also analyzed by electrophoresis on 2% agarose gels, and gel images were captured with a G:BOX gel imaging system (Syngene).
2.4. Immunocytochemistry
Cells were seeded on sterile 13‐mm circular glass coverslips placed in 24‐well plates at 20 000 cells per well and incubated for 48 hours. Cells were washed twice with PBS and fixed with 4% paraformaldehyde for 20 minutes. After washed with PBS three times, cells were permeabilized by incubation in PBS containing 0.3% Triton X‐100 for 5 minutes. Cells were washed three times with PBS and incubated in PBS containing 10% goat serum for 1 hour at room temperature. Cells were incubated with the primary rabbit anti‐Piezo1 antibody (Proteintech) at a dilution of 1:100 overnight at 4°C. After washed with PBS three times, cells were incubated with the secondary fluorescein isothiocyanate‐conjugated goat anti‐rabbit IgG antibody at 1:500 at room temperature for 1 hour in dark. After washed with PBS and rinsed with water, cells were mounted on a microscope slide with antifade mounting medium containing 4′,6‐diamidino‐2‐phenylindole (DAPI) (Invitrogen). Fluorescent images were captured using a LMS880 confocal microscope and the ZEN software (Zeiss) or an EVOS Cell Imaging System (Thermo Fisher Scientific, Waltham, MA, USA).
2.5. Measurements of intracellular Ca2+ concentration
The intracellular Ca2+ concentration ([Ca2+]i) was monitored using Fura‐2/AM and a Flex‐Station III microplate reader (Molecular Devices) as described in our previous study.40 Cells were seeded at 40 000 cells per well in 96‐well microplates (SARSTEDT) and incubated for 24 hours. Cells were washed with Ca2+‐containing standard buffer solution (SBS in mM: NaCl 134, KCl 5, CaCl2 1.5, MgCl2 1.2, HEPES 2.38 and glucose 8, pH 7.4) and incubated with SBS containing 4 μM Fura‐2/AM and 0.04% pluronic acid F‐127 at 37°C for 45 minutes in dark. Cells were washed with SBS twice and incubated for 30 minutes with 160 μL of Ca2+‐containing or Ca2+‐free SBS (in mM: NaCl 134, KCl 5, MgCl2 1.2, HEPES 2.38, glucose 8 and EGTA 0.4, pH 7.4) to investigate Yoda1‐induced change in the [Ca2+]i. After the basal line was established for 30‐60 seconds, 40 μL of SBS containing Yoda1 at five times of the indicated final concentrations was transferred into each well from the compound plate into the cell plate. In the experiments studying the effects of RR or GsMTx4 on Yoda1‐induced increase in the [Ca2+]i, cells were treated with RR for 5 minutes during the recording or with GsMTx4 30 minutes prior to and during the recording. The ratio of fluorescence intensity, excited at 340 and 380 nm, respectively, and emitted at 510 nm (F 340/F 380), was used to indicate the [Ca2+]i. Data analysis was carried out using Origin. The mean data are presented by expressing the F 340/F 380 as a percentage of the F 340/F 380 from cells under the control conditions.
2.6. Transfection with siRNA
Cells were seeded in 96‐well microplates or 24‐well plates at 40 000 cells per well for measurement of the [Ca2+]i and migration assay, respectively, and in 6‐well plates at 80000 cells per well for RT‐qPCR. After 24 hours, cells were transfected with 30‐50 nM of Piezo1‐specific siRNA (siPiezo1) and control siRNA (siCTL) (control siRNA1#; Ambion or Dharmacon), using Lipofectamine RNAiMAX transfection reagent (Invitrogen) according to the manufacturer's instructions. Cells were used for measurement of the [Ca2+]i and migration assays 48 hours and for RT‐qPCR 72 hours posttransfection. The difference in the Piezo1 mRNA expression in cells transfected with siPiezo1 relative to that in cells transfected with siCTL was calculated using the 2‐ΔΔCt method, where ΔΔCt = [(CtPiezo1 − Ctβ‐actin)siPiezo1 − (CtPiezo1 − Ctβ‐actin)siCTL].72
2.7. Cell migration assay
Cell migration was determined using the wound healing assay as described in our previous study,40 and the wound area was created using a two‐chamber insert (Ibidi) placed in the middle of each well of 24‐well plates. Cells were seeded at 20 000‐25 000 cells per chamber and incubated for 16‐17 hours. The insert was lifted carefully, and cells were gently rinsed with PBS. Fresh culture media supplemented with 2% FBS with or without containing Yoda1 were added into each well. In experiments examining the effects of inhibitors (apyrase, PPADS, PF431396, and U0126) on cell migration, cells were pretreated with the inhibitor at the indicated concentrations for 30 minutes and the inhibitor continued to be present during the whole essay. Phase contrast images were captured using an EVOS Cell Imaging System at 0, 24, 48, and 72 hours. The wound area was calculated using the TScratch software. For individual experiments, wound healing was derived by the difference between the initial wound area and the ones at 24, 48, or 72 hours and expressed as percentage of the initial wound area (eg, Figure 3B‐E). To mitigate the variations between different experiments, the mean would healing was presented by expressing the wound healing as percentage of that under control condition at the same time points (eg, Figure 3F).
2.8. Measurement of ATP release
ATP release was determined using a StayBrite Highly Stable ATP Bioluminescence Assay kit (BioVision) to measure the ATP concentration in the culture medium according to the manufacturer's instructions. Cells were plated at 50 000‐60 000 cells/well in 200 μL of standard culture medium in a 96‐well plate and incubated overnight. Next day, cells were subjected to starvation by replacing the culture medium with fresh media without FBS and incubating for 2 hours. Cells were treated with 100 μM suramin for 30 minutes at 37°C in order to prevent ATP hydrolysis, as described in a previous study.38 After cells were exposed to 1 μM Yoda1 for 30 minutes at 37°C, the culture medium were collected, placed in 1.5‐mL Eppendorf tubes on ice, and centrifuged at 11 000g for 5 minutes at 4°C. The supernatants were transferred to a 96‐well plate with 10 μL per well in triplicate for each condition. The luminescence intensity was measured using a Flex‐Station III microplate reader. The ATP concentration was derived using a standard curve constructed using 10, 30, 100, 1000, and 10 000 nM ATP.
2.9. Data presentation and statistical analysis
All data are presented as mean ± SEM, where appropriate. Statistical analysis was performed using Origin; Student's t test was used for comparison between two groups, and one‐way ANOVA followed by Fisher's test for comparison of multiple groups, with P < .05 being statistically significant.
3. RESULTS
3.1. The Ca2+‐permeable piezo1 channel is expressed in hDP‐MSCs
We began with using RT‐qPCR to analyze the Piezo1 expression. The Piezo1 mRNA was readily detected in hDP‐MSCs from all four donors, albeit with some variations in the mRNA level among the different donors (Figure 1A and Supplementary Figure S1). Consistently, as observed in cells from all the donors, there were positive and also variable immunoreactivities in cells labeled with the anti‐Piezo1 antibody but not in cells labeled only with the secondary antibody (Figure 1B). We next examined the functional expression of the Piezo1 channel in hDP‐MSCs by monitoring intracellular Ca2+ responses to Yoda1, the Piezo1 channel‐specific activator.73 In extracellular Ca2+‐containing solution, brief exposure to Yoda1 (0.1‐10 μM) induced concentration‐dependent Ca2+ responses with ≥1 μM Yoda1 inducing a significant increase in the [Ca2+]i in cells from two donors examined (Figure 1C,D). In hDP‐MSCs from all four donors, brief exposure to 3 μM Yoda1 consistently evoked robust increases in the [Ca2+]i in extracellular Ca2+‐containing solution and, by contrast, no or very little change in the [Ca2+]i in extracellular Ca2+‐free solution (Figure 1E‐H), indicating that Yoda1 almost exclusively induced extracellular Ca2+ influx. Next, we determined the effects of RR, which is known to inhibit the Piezo1 channel with a potency of ~5 μM,43 and GsMTx4, a peptide from tarantula venom that inhibits the Piezo1 channel in the low μM concentrations,74 on Yoda1‐induced Ca2+ responses. Treatment with 10 or 30 μM RR for 5 minutes exerted no effect on the basal Ca2+ level but strongly and concentration‐dependently reduced Yoda1‐induced increase in the [Ca2+]i in cells from three donors examined (Figure 2A‐D and Supplementary Figure S2). Treatment with 0.3, 1 or 3 μM GsMTx4 for 30 minutes also resulted in a concentration‐dependent inhibition of Yoda1‐induced increase in the [Ca2+]i in cells from two donors examined (Figure 2E‐G and Supplementary Figure S3). Finally, we examined the effect of siRNA‐mediated knockdown of the Piezo1 expression on Yoda1‐induced Ca2+ responses. Genetic depletion of the Piezo1 expression (Figure 2H) strongly suppressed Yoda1‐induced increase in the [Ca2+]i in cells from three donors tested (Figure 2I‐L and Supplementary Figure S4). Taken together, these results provide pharmacological and genetic evidence that consistently supports the expression of the Piezo1 channel in hDP‐MSCs.
Figure 1.
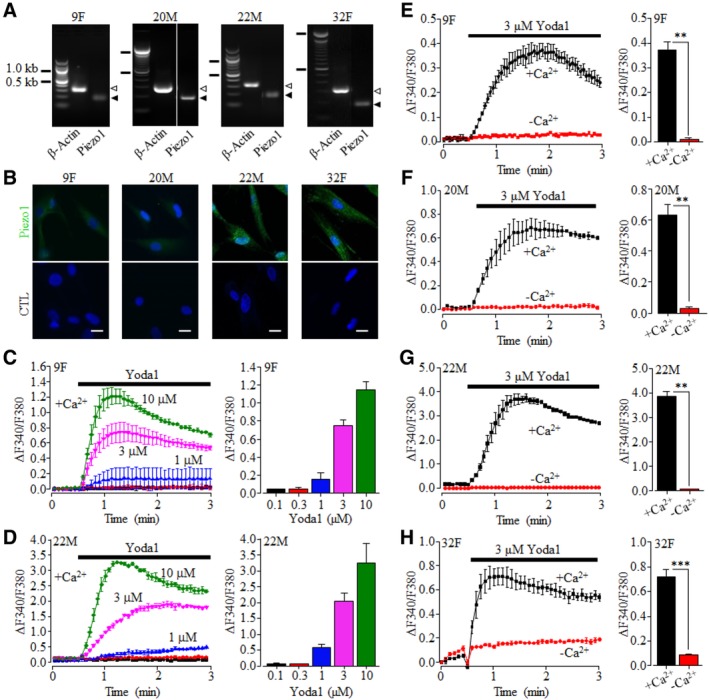
Expression of the Ca2+‐permeable Piezo1 channel on hDP‐MSC. A, Representative agarose gel images showing positive detection by RT‐PCR of Piezo1 mRNA expression in hDP‐MSC cells from each of the four donors examined (9F, 20M, 22M, and 32F). Similar results were observed in at least two independent experiments. The solid and hollow arrowheads on the right indicate the PCR products for β‐actin and Piezo1, respectively. B, Representative immunofluorescent images showing staining of cells from each of the four donors with the anti‐Piezo1 antibody (top) or only with the second antibody (bottom). Scale bar = 20 μm. C,D, Representative intracellular Ca2+ responses in Ca2+‐containing solution (+Ca2+) to 0.1 (black), 0.3 (red), 1 (blue), 3 (magenta), and 10 μM (green) Yoda1 (left) and mean peak Ca2+ responses (right) from one set of experiments using four wells of cells for each concentration, in cells from 9F (C) and 22M (D). E‐H, Representative Ca2+ responses to 3 μM Yoda1 in Ca2+‐containing (+Ca2+) or Ca2+‐free (‐Ca2+) solutions in parallel experiments (left) and mean peak Ca2+ responses (right) from one set of experiment using four wells of cells for each condition, in cells from 9F (E), 20M (F), 22M (G), and 32F (H). **P < .01 and ***P < .001
Figure 2.
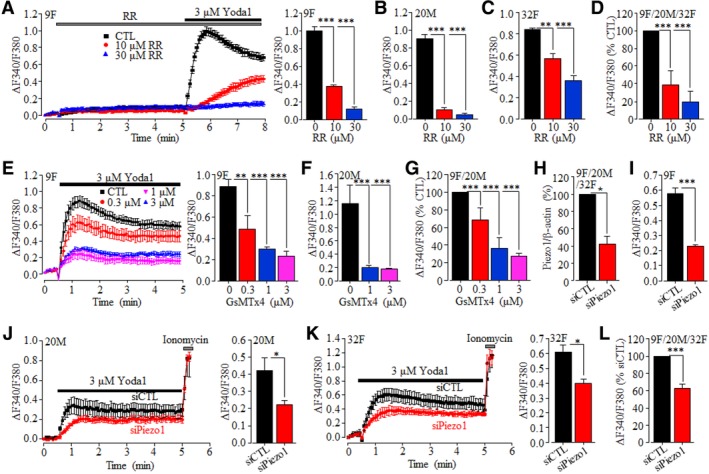
Pharmacological and genetic inhibition of Yoda1‐induced Piezo1‐mediated intracellular Ca2+ responses in hDP‐MSC. A‐C, Yoda1‐induced Ca2+ responses (9F) and peak Ca2+ responses (9F, 20M, and 32F) in control cells and cells treated with ruthenium red (RR) for 5 minutes from one set of experiments using 3‐4 wells of cells for each condition. D, Summary of Yoda1‐induced peak Ca2+ responses in RR‐treated cells from 9F, 20M, and 32F, as percentage of that in control cells, from three independent experiments. E,F, Yoda1‐induced Ca2+ responses (9F) and peak Ca2+ responses (9F and 20M) in control and cell prior treated with GsMTx4 for 30 minutes from one set of experiments using 3‐4 wells of cells for each condition. G, Summary of Yoda1‐induced peak Ca2+ responses in GsMTx4‐treated cells as percentage of that in control cells from four independent experiments using cells from 9F and 20M. H, Summary of Piezo1/β‐actin in cells transfected with siCTL or siPiezo1 as percentage of that in siCTL‐transfected cells from eight independent experiments using cells from 9F, 20M, and 32F. I‐K, Yoda1‐induced Ca2+ responses in cells transfected with siCTL or siPiezo1 from 9F (I), 20M (J), and 32F (K) from one set of experiments using four wells of cells for each condition. Cells were exposed to 5 μM ionomycin at the end of recordings (J, K). L, Summary of peak Yoda1‐induced Ca2+ responses in siPiezo1‐transfected cells as percentage of that in siCTL‐transfected cells from five independent experiments using from 9F, 20M, and 32F, *P < .05; **P < .01; and ***P < .001
3.2. Activation of the Piezo1 channel stimulates hDP‐MSC migration
To test whether the Piezo1 channel plays a role in regulating hDP‐MSC migration, as recently reported in endothelial cell56 and various cancer cells,50, 57, 58, 59 we used the wound healing assay to examine the effect of Yoda1 on cell migration (Figure 3). Inclusion of 0.1‐1 μM Yoda1 in the culture medium resulted in a concentration‐dependent increase in cell migration, with 0.3 and 1 μM Yoda1 significantly accelerating cell migration at 24, 48, and 72 hours, and such stimulation was observed in hDP‐MSCs from all four donors, with some variations among the different donors (Figure 3A‐F). Prolonged exposure to Yoda1 at 3 μM and particularly 10 μM for 24‐72 hours was detrimental to hDP‐MSCs (data not shown). Unfortunately, prolonged exposure to RR or GsMTx4 was also cytotoxic, preventing from pharmacologically interrogating the role of the Pieoz1 channel in mediating Yoda1‐induced stimulation of cell migration. We therefore turned to examine the effect of siRNA‐mediated knockdown of the Piezo1 expression on cell migration. Yoda1‐induced stimulation of cell migration was sustained in cells transfected with control siRNA but lost in cells transfected with Piezo1‐specific siRNA, which was consistently observed in cells from two donors tested (Figure 3G). These results support that activation of the Piezo1 channel stimulates hDP‐MSC migration.
Figure 3.
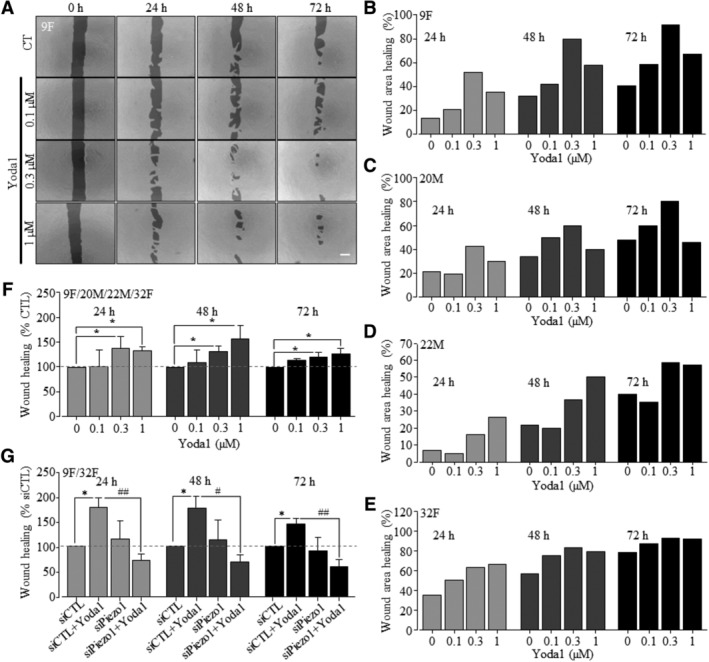
Yoda1‐induced activation of the Piezo1 channel stimulates hDP‐MSC migration. A, Representative images showing the wound areas at 0, 24, 48, and 72 hours, using cells from 9F in the absence (CTL) and presence of indicated concentrations of Yoda1 in the culture medium. Scale bar = 500 μm. B‐E, Wound healing indicated by the reduction in the wound areas as percentage of the wound area at 0 hour using cells from 9F (B, as shown in A), 20M (C), 22M (D), and 32F (E). F, Summary of the mean wound healing as percentage of that under control conditions at the same time points in parallel experiments from eight independent experiments using all four donors shown in A‐E. G, Summary of mean wound healing under indicated conditions, from five independent experiments using cells from 9F and 32F that were transfected with control siRNA (siCTL) and Piezo1‐specific siRNA (siPiezo1). *P < .05 compared to control condition at the same time points. # P < .05 and ## P < .001 compared to cells transfected with siCTL and exposed to Yoda1
3.3. PYK2 and MEK/ERK are engaged in Yoda1‐induced stimulation of hDP‐MSC migration
As introduced above, we were interested in examining whether the PYK2 and MEK/ERK signaling pathways participate in Piezo1‐dependent regulation of hDP‐MSC migration. In hDP‐MSCs from three donors examined, treatment with 10 nM PF431396, a PYK2 inhibitor, prior to and during exposure to Yoda1, strongly inhibited Yoda1‐induced stimulation of cell migration (Figure 4A‐D). Similarly, treatment with 1 μM U0126, a MEK/ERK inhibitor, resulted in strong inhibition of Yoda1‐induced stimulation of cell migration (Figure 4E‐H). These results provide clear evidence to suggest that the PYK2 and MEK/ERK signaling pathways are critically engaged in Piezo1‐dependent regulation of hDP‐MSC migration.
Figure 4.
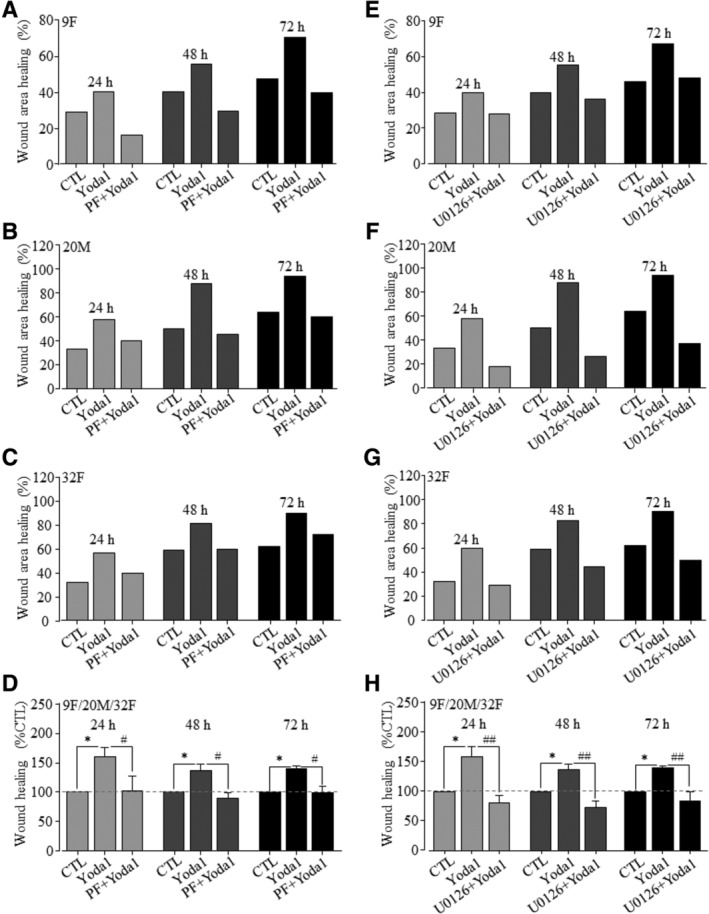
The PYK2 and MEK/ERK signaling pathways are critical for Yoda1‐induced stimulation of hDP‐MSC migration. A‐C, Wound healing for cells from 9F (A), 20M (B), and 32F (C) under control condition (CTL) and cells exposed to 0.3 μM Yoda1 alone (Yoda1) or together with 10 nM PF431396 (PF + Yoda1). D, Summary of mean wound healing in three independent experiments for cells from 9F, 20M, and 32F under control condition (CTL) and cells exposed to Yoda1 alone or together with PF as shown in A‐C. E‐G, Wound healing for cells from 9F (E), 20M (F), and 32F (G) under CTL and cells exposed to 0.3 μM Yoda1 alone (Yoda1) or together with 1 μM U0126 (U0126 + Yoda1). H, Summary of mean wound healing in three independent experiments for cells from 9F, 20M, and 32F under control condition (CTL) and cells exposed to Yoda1 alone or together with U0126 as shown in E‐G. *P < .05 compared to control condition at the same time points, and # P < .05 and ## P < .001 compared to cells exposed to Yoda1
3.4. Yoda1‐induced hDP‐MSC migration depends on ATP release and P2 receptor activation
Next, we examined the hypothesis that activation of the Piezo1 channel stimulates hDP‐MSC migration via inducing ATP release and subsequent activation of the P2 receptor purinergic signaling. We examined Yoda1‐induced ATP release from hDP‐MSCs (Figure 5A). Inclusion of 100 μM suramin in the culture medium to prevent ATP hydrolysis38 resulted in slight but statistically insignificant elevation in the ATP concentration, consistent with a previous study suggesting a low level of ATP release under the normal culturing conditions. 36 Exposure to 1 μM Yoda1 for 30 minutes resulted in a 5.5‐fold increase in the ATP concentration (Figure 5A), indicating that activation of the Piezo1 channel induces ATP release. We moved on to determine the effect of treatment with apyrase, an ATP‐scavenging enzyme, on Yoda1‐induced increase in hDP‐MSC migration. Inclusion of 1 U/mL apyrase in the culture medium completely prevented Yoda1‐induced stimulation of cell migration. Treatment with 0.3 U/mL apyrase also significantly reduced Yoda1‐induced increase in cell migration at 24 hours but became less effective at 48 hours or ineffective at 72 hours (Figure 5B). Importantly, treatment with apyrase alone was without significant effect on cell migration (Supplementary Figure S5A). These results indicate that ATP release mediates Yoda1‐induced stimulation of hDP‐MSC migration. Our previous study showed that treatment with 30 μM PPADS, a generic P2 receptor antagonist, prior to and during exposure to ATP, effectively prevented ATP‐induced stimulation of hDP‐MSC migration.40 Treatment with 30 μM PPADS also resulted in strong inhibition of Yoda1‐induced stimulation of cell migration (Figure 5C). Again, treatment with PPADS alone had no effect on cell migration (Supplementary Figure S5B). Taken together, these results provide consistent evidence to show that activation of the Piezo1 channel stimulates hDP‐MSC migration via inducing ATP release and P2 receptor activation.
Figure 5.
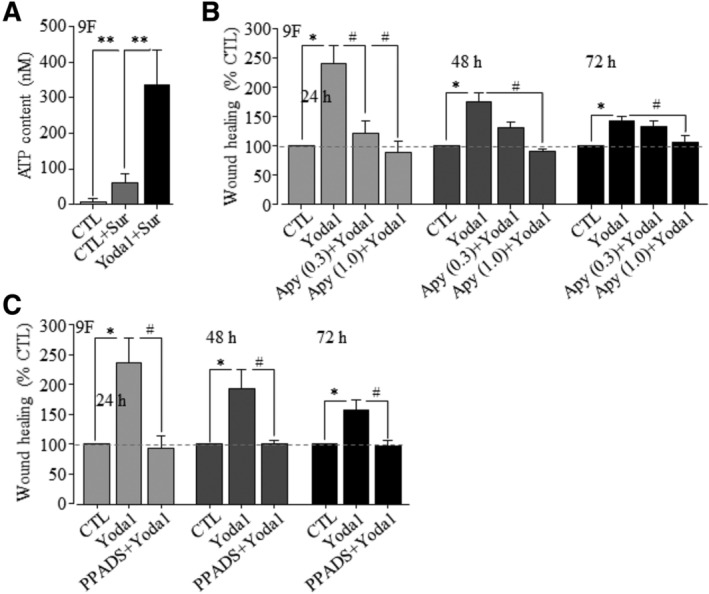
Yoda1‐induced stimulation of hDP‐MSC migration depends ATP release and activation of P2 purinergic signaling. A, The ATP concentrations in the medium culturing cells under control condition (CTL) or treated with 100 μM suramin (CTRL+Sur) or with 100 μM suramin and 1 μM Yoda1 (Yoda1 + Sur). The results were obtained in triplicates. **P < .01. B, Summary of mean wound healing from three independent experiments for cells from 9F under control condition (CTL) and cells exposed to 0.3 μM Yoda alone (Yoda1) or together with 0.3 or 1 U/mL apyrase (Apy + Yoda1). C, Summary of mean wound healing in three independent experiments for cells from 9F under CTL and cells exposed to 0.3 μM Yoda1 alone (Yoda1) or together with 30 μM PPADS (PPAD+Yoda1). *P < .05 compared to control condition at the same time points and # P < .05 compared to cells exposed to Yoda1 alone
If the above‐described notion is correct, we anticipate that activation of the PKY2 and MEK/ERK signaling pathways are also critical in ATP‐induced stimulation of hDP‐MSC migration. Exposure to 30 μM ATP stimulated cell migration in hDP‐MSCs from two donors used (Figure 6A,B), as reported in our previous study.40 Indeed, ATP‐induced stimulation of cell migration was prevented by treatment with PF431396 or U0126 (Figure 6C,D), as shown above for Yoda1 (Figure 4). These results have extended our previous finding40 to show engagement of the PYK2 and MEK/ERK signaling pathways in ATP‐induced stimulation of hDP‐MSC migration. Such results are highly consistent with the notion that activation of the Piezo1 channel stimulates hDP‐MSC migration via inducing ATP release and subsequent activation of the P2 receptor purinergic signaling and downstream PYK2 and MEK/ERK signaling pathways.
Figure 6.
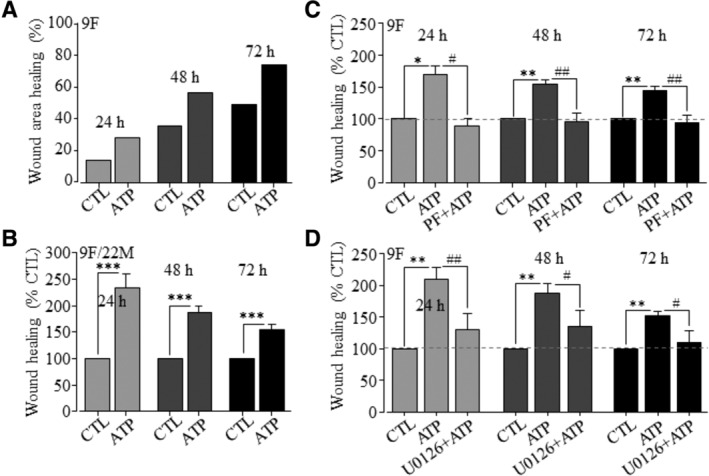
The PYK2 and MEK/ERK signaling pathways are critical for ATP‐induced stimulation of hDP‐MSC migration. A, Representative wound healing in cells from 9F under control (CTL) or cells exposed to 30 μM ATP, indicated by the reduction in the wound area expressed as percentage of the wound area at 0 hour. B, Summary of mean wound healing as percentage of that under CTL at the same time points in parallel experiments, from 10 independent experiments using cells from 9F and 22M. C,D, Summary of mean wound healing in three independent experiments for cells from 9F under CTL and cells exposed to 30 μM ATP alone (ATP) or together with 10 nM of PF431396 (PF + ATP, C) or 1 μM U0126 (U0126 + ATP, D). *P < .05 and **P < .001 compared to control condition at the same time points, and # P < .05 and ## P < .001 compared to cells exposed to ATP alone (C and D)
4. DISCUSSION
We in this study showed the expression of the Piezo1 channel in hDP‐MSCs as a previously unrecognized mechanism regulating human MSC migration. We provide further evidence to suggest that activation of the Piezo1 channel regulates cell migration via inducing ATP release and subsequent activation of the P2 receptor purinergic signaling and downstream PYK2 and MEK/ERK signaling pathways (Figure 7). These novel findings provide a better understanding of the mechanisms mediating ATP release from MSCs and regulation of MSC migration. Such information is useful in developing strategies to improve MSC homing and applications of MSCs in regenerative medicine and tissue engineering.
Figure 7.
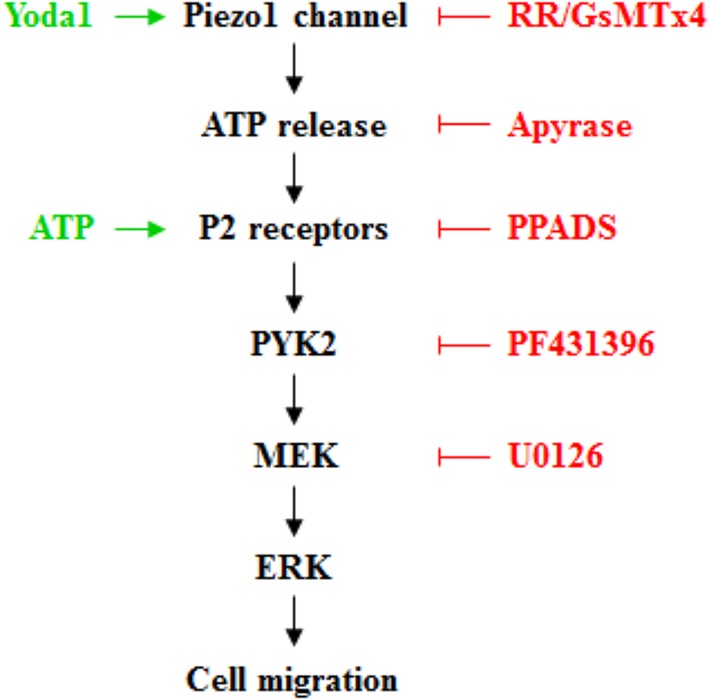
Schematic summary of proposed signaling mechanisms for Piezo1‐dependent ATP release from hDP‐MSC and regulation of cell migration. Activation of the Piezo1 channel induces ATP release. ATP activates the P2 receptors and induces activation of the proline‐rich tyrosine kinase 2 (PYK2) and the mitogen‐activated protein kinase/extracellular signal‐regulated kinase (MEK/ERK), which may act as upstream and downstream signaling pathways (as depicted here) or independent signaling pathways, leading to an increase in cell migration. The activators and inhibitors used in this study to target various signaling molecules are highlighted in green and red, respectively
Our study provides independent lines of evidence to demonstrate the expression of the Piezo1 channel in hDP‐MSCs. The Piezo1 mRNA and protein were detected in all the cell preparations from four donors, albeit with some noticeable variations among the different donors (Figure 1A,B and Supplementary Figure S1). Consistently, brief exposure to Yoda1 induced Ca2+ influx‐dependent increases in the [Ca2+]i in hDP‐MSCs from all the donors (Figure 1C‐H). Moreover, as shown in cells from multiple donors, Yoda1‐induced Ca2+ responses were inhibited by RR and GsMTx4, and also by siRNA‐mediated knockdown of the Piezo1 expression (Figure 2 and Supplementary Figures S2‐S4). These results provide strong evidence to show the expression of the Piezo1 channel in hDP‐MSCs. Our study, together with a recent study examining hBM‐MSCs,60 supports the expression of the Piezo1 channel in human MSCs of different tissues. Yoda1‐induced increase in the [Ca2+]i in hDP‐MSCs was abolished by removing extracellular Ca2+ (Figure 1E‐H), suggesting that the Piezo1 channel is located in the plasma membrane, as described in many other types of cells.43, 44, 54
Recent studies have reported a role for the Piezo1 channel in regulating cell migration in endothelial cell56 and cancer cells.50, 57, 58, 59 In this study, we provide evidence to show that the Piezo1 channel plays a similar role in hDP‐MSCs. Persistent exposure to Yoda1 stimulated hDP‐MSC migration, and such stimulatory effect was consistently observed in cells from all the four donors (Figure 3A‐F). Unfortunately, prolonged treatment with RR or GsMTx4 caused cytotoxicity to hDP‐MSCs, thus preventing from using them as pharmacological means to examine the role of the Piezo1 channel in Yoda1‐induced stimulation of cell migration. Nonetheless, we showed that siRNA‐mediated knockdown of the Piezo1 expression strongly inhibited Yoda1‐induced stimulation of cell migration (Figure 3G), supporting a significant role for the Piezo1 channel in regulating hDP‐MSC migration.
In this study, we have gained further insights into Piezo1‐dependent regulation of hDP‐MSC migration. Recent studies support a critical role for the Piezo1 channel, upon mechanical or chemical activation, in inducing ATP release from urothelial cell,54 red blood cell,55 and endothelial cell.48 Here, we showed that activation of the Piezo1 channel with Yoda1 induced substantial ATP release from hDP‐MSCs (Figure 5A). Moreover, Yoda1 became ineffective in stimulating cell migration in the presence of apyrase, a potent ATP scavenger (Figure 5B), indicating that ATP release mediates Piezo1‐dependent regulation of cell migration. These findings are nicely aligned with our previous study showing that application of ATP stimulates hDP‐MSC migration under the same experimental conditions,40 which we have confirmed in this study (Figure 6A,B). We further showed that Yoda1‐induced Piezo1‐dependent stimulation of hDP‐MSC migration was strongly inhibited by PPADS (Figure 5B). Taken together, these results strongly support that activation of the Piezo1 channel stimulates hDP‐MSC migration via inducing ATP release and subsequent activation of the P2 receptor purinergic signaling. Our previous study identified P2X7, P2Y1, and P2Y11 as the major P2 receptors that mediate ATP‐induced increase in hDP‐MSC migration.40 It is anticipated that these P2 receptors are engaged in Piezo1‐dependent stimulation of hDP‐MSC migration but the supporting evidence is required.
In this study, we have also shed light on the downstream signaling pathways in Piezo1‐dependent stimulation of hDP‐MSC migration as well as ATP‐induced P2 receptor‐dependent stimulation of hDP‐MSC migration reported in our previous study.40 PYK2 is a well‐recognized signaling mechanism transducing Ca2+‐dependent induction of the MEK/ERK signaling pathway.64, 66, 67, 69, 70 Our results from hDP‐MSCs from three donors showed that Yoda1‐induced cell migration was prevented by inhibiting PYK2 (Figure 4A‐D) and MEK/ERK (Figure 4E‐H). These results strongly suggest that the PYK2 and MEK/ERK signaling pathways are critically engaged in Piezo1‐dependent stimulation of cell migration. Our finding is also consistent with a recent study proposing a role for the Piezo1 channel in rat DP‐MSCs in mediating ultrasound‐induced activation of the MEK/ERK signaling pathway.61 ATP‐induced P2 receptor‐dependent stimulation of hDP‐MSC migration was also prevented by inhibiting PYK2 and MEK/ERK (Figure 6C,D). Such findings are interesting as they are clearly consistent with the idea that activation of the Piezo1 channel stimulates cell migration via inducing ATP release and subsequent activation of the P2 receptor purinergic signaling and downstream PYK2 and MEK/ERK signaling pathways (Figure 7). There is evidence that mechanical stimulus‐induced increase in the [Ca2+]i in MSCs and other cell types depends on release of ATP and subsequent activation of the P2 receptor purinergic signaling.35, 75 It is interesting to establish whether mechanical activation of the Peizo1 channel can induce ATP release and regulate hDP‐MSC migration, as we have shown here for chemical activation of the Pieoz1 channel. Moreover, both ATP‐induced activation of the P2 purinergic signaling reported in our previous study40 and activation of the Piezo1 channel as shown in the present study can elevate the [Ca2+]i. Further investigations are required to better understand how the Ca2+ signals from these two distinctive mechanisms regulate hDP‐MSC migration.
5. CONCLUSION
In conclusion, we show that activation of the Piezo1 channel as a novel mechanism to stimulate in MSC migration via inducing ATP release and subsequent activation of the P2 receptor purinergic signaling and downstream PYK2 and MEK/ERK signaling pathways. Such novel insights into the molecular and signaling mechanisms in the regulation of MSC migration should be useful for a better understanding of the MSC biology and translational use of MSCs in regenerative medicine and tissue engineering.
CONFLICT OF INTEREST
The authors indicated no potential conflicts of interest.
AUTHOR CONTRIBUTIONS
L.‐H.J: conception and design of the research, interpretation of data, and manuscript writing and revision; H.P.: conception and design of the research and interpretation of data; F.M.: conception and design of the research, and interpretation of data, collection and analysis of data, manuscript writing and revision, and collection and analysis of data; J.L., P.S., S.R., H.Z., X.Y.: intellectual inputs, provision of materials, and interpretation of data. All authors provided critical comments and approved the manuscript.
Supporting information
Figure 1. RT‐qPCR analysis of Piezo1 mRNA expression in human DP‐MSCs.
Representative RT‐qPCR amplification plots (left) and mean Piezo1/β‐actin (right) from 3 independent mRNA preparations for 9F, and 2 independent mRNA preparations for 22M, 20M and 32F cells.
Figure 2. Effects of ruthenium red on Yoda1‐induced Ca 2+ responses in human DP‐MSCs.
Representative Yoda1‐induced intracellular Ca2+ responses to 3 μM Yoda1 in cells from 20M (A) or 32F (B) without (CTL) or after treatment with 10 or 30 μM ruthenium red (RR) for 5 minutes from one set of experiments using 4 wells of cells for each condition.
Figure 3. Effect of GsMTx4 on Yoda1‐induced Ca 2+ responses in human DP‐MSCs.
Representative Yoda1‐induced intracellular Ca2+ responses to 3 μM Yoda1 in cells from 20M without (CTL) and with treatment with 1 or 3 μM GsMTx4 for 30 minutes prior and during exposure to Yoda1 from one set of experiments using 4 wells of cells for each condition.
Figure 4. Effect of siRNA‐mediated knockdown of Piezo1 expression on Yoda1‐induced Ca 2+ response in hDP‐MSCs
Representative intracellular Ca2+ responses to 3 μM Yoda1 from one set of experiments using 4 wells of cells for each condition, in cells from 9F that were transfected with control siRNA (siCTL) or Piezo1‐specific siRNA (siPiezo1). Cells were exposed to 5 μM ionomycin at the end of recordings.
Figure 5. No effect of apyrase or PPADS on human DP‐MSC migration.
Summary of mean wound narrowing in 3 independent experiments for cells from 9F under control condition (CTL) and cells exposed to 0.3 or 1 U/mL apyrase (Apy) (A) or 30 μM PPADS in cells from 9F and 22M (B). NS, no significant difference.
ACKNOWLEDGMENTS
We are grateful to the persons who kindly donated their teeth for cell preparations used in this study. Fatema Mousawi was supported by a PhD scholarship from Kuwait High Commission and Hongsen Peng by a PhD scholarship from University of Leeds.
Mousawi F, Peng H, Li J, et al. Chemical activation of the Piezo1 channel drives mesenchymal stem cell migration via inducing ATP release and activation of P2 receptor purinergic signaling. Stem Cells. 2020;38:410–421. 10.1002/stem.3114
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding author upon reasonable request.
REFERENCES
- 1. Dominici M, Le Blanc K, Mueller I, et al. Minimal criteria for defining multipotent mesenchymal stromal cells. The International Society for Cellular Therapy position statement. Cytotherapy. 2006;8:315‐317. [DOI] [PubMed] [Google Scholar]
- 2. Chen Q, Shou P, Zheng C, et al. Fate decision of mesenchymal stem cells: adipocytes or osteoblasts? Cell Death Differ. 2016;23:1128‐1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Liu ZJ, Zhuge Y, Velazquez OC. Trafficking and differentiation of mesenchymal stem cells. J Cell Biochem. 2009;106:984‐991. [DOI] [PubMed] [Google Scholar]
- 4. Volkman R, Offen D. Concise review: mesenchymal stem cells in neurodegenerative diseases. Stem Cells. 2017;35:1867‐1880. [DOI] [PubMed] [Google Scholar]
- 5. Nitkin CR, Bonfield TL. Concise review: mesenchymal stem cell therapy for pediatric disease: perspectives on success and potential improvements. Stem Cells Translational Medicine. 2017;6:539‐565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bhat IA, Sivanarayanan TB, Somal A, et al. An allogenic therapeutic strategy for canine spinal cord injury using mesenchymal stem cells. J Cell Physiol. 2019;234:2705‐2718. [DOI] [PubMed] [Google Scholar]
- 7. Wang YH, Wu DB, Chen B, Chen EQ, Tang H. Progress in mesenchymal stem cell‐based therapy for acute liver failure. Stem Cell Res Ther. 2018;9:227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Volarevic V, Nurkovic J, Arsenijevic N, Stojkovic M. Concise review: therapeutic potential of mesenchymal stem cells for the treatment of acute liver failure and cirrhosis. Stem Cells. 2014;32:2818‐2823. [DOI] [PubMed] [Google Scholar]
- 9. Marcheque J, Bussolati B, Csete M, Perin L. Concise reviews: stem cells and kidney regeneration: an update. Stem Cells Translational Medicine. 2019;8:82‐92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Peltzer J, Aletti M, Frescaline N, Busson E, Lataillade JJ, Martinaud C. Mesenchymal stromal cells based therapy in systemic sclerosis: rational and challenges. Front Immunol. 2018;9:2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Marofi F, Vahedi G, Hasanzadeh A, et al. Mesenchymal stem cells as the game‐changing tools in the treatment of various organs disorders: mirage or reality? J Cell Physiol. 2019;234:1268‐1288. [DOI] [PubMed] [Google Scholar]
- 12. Bagheri‐Mohammadi S, Karimian M, Alani B, Verdi J, Tehrani RM, Noureddini M. Stem cell‐based therapy for Parkinson's disease with a focus on human endometrium‐derived mesenchymal stem cells. J Cell Physiol. 2019;234:1326‐1335. [DOI] [PubMed] [Google Scholar]
- 13. Galipeau J, Sensebe L. Mesenchymal stromal cells: clinical challenges and therapeutic opportunities. Cell Stem Cell. 2018;22:824‐833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ward MR, Abadeh A, Connelly KA. Concise review: rational use of mesenchymal stem cells in the treatment of ischemic heart disease. Stem Cells Translational Medicine. 2018;7:543‐550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Loebel C, Burdick JA. Engineering stem and stromal cell therapies for musculoskeletal tissue repair. Cell Stem Cell. 2018;22:325‐339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. McGonagle D, Baboolal TG, Jones E. Native joint‐resident mesenchymal stem cells for cartilage repair in osteoarthritis. Nat Rev Rheumatol. 2017;13:719‐730. [DOI] [PubMed] [Google Scholar]
- 17. Scolding NJ, Pasquini M, Reingold SC, et al. Cell‐based therapeutic strategies for multiple sclerosis. Brain. 2017;140:2776‐2796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lo Furno D, Mannino G, Giuffrida R. Functional role of mesenchymal stem cells in the treatment of chronic neurodegenerative diseases. J Cell Physiol. 2018;233:3982‐3999. [DOI] [PubMed] [Google Scholar]
- 19. Laroye C, Gibot S, Reppel L, Bensoussan D. Concise review: mesenchymal stromal/stem cells: a new treatment for sepsis and septic shock? Stem Cells. 2017;35:2331‐2339. [DOI] [PubMed] [Google Scholar]
- 20. Majka M, Sulkowski M, Badyra B, et al. Concise review: mesenchymal stem cells in cardiovascular regeneration: emerging research directions and clinical applications. Stem Cells Translational Medicine. 2017;6:1859‐1867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Higuchi A, Kumar SS, Benelli G, et al. Stem cell therapies for reversing vision loss. Trends Biotechnol. 2017;35:1102‐1117. [DOI] [PubMed] [Google Scholar]
- 22. Maijenburg MW, van der Schoot CE, Voermans C. Mesenchymal stromal cell migration: possibilities to improve cellular therapy. Stem Cells Dev. 2012;21:19‐29. [DOI] [PubMed] [Google Scholar]
- 23. Sohni A, Verfaillie CM. Mesenchymal stem cells migration homing and tracking. Stem Cells Int. 2013;2013:130763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. De Becker A, Riet IV. Homing and migration of mesenchymal stromal cells: how to improve the efficacy of cell therapy? World J Stem Cells. 2016;8:73‐87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Nitzsche F, Muller C, Lukomska B, et al. Concise review: MSC adhesion cascade‐insights into homing and transendothelial migration. Stem Cells. 2017;35:1446‐1460. [DOI] [PubMed] [Google Scholar]
- 26. Rombouts WJ, Ploemacher RE. Primary murine MSC show highly efficient homing to the bone marrow but lose homing ability following culture. Leukemia. 2003;17:160‐170. [DOI] [PubMed] [Google Scholar]
- 27. Sipp D, Robey PG, Turner L. Clear up this stem‐cell mess. Nature. 2018;561:455‐457. [DOI] [PubMed] [Google Scholar]
- 28. Samsonraj RM, Raghunath M, Nurcombe V, Hui JH, van Wijnen AJ, Cool SM. Concise review: multifaceted characterization of human mesenchymal stem cells for use in regenerative medicine. Stem Cells Translational Medicine. 2017;6:2173‐2185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Schwab A, Fabian A, Hanley PJ, Stock C. Role of ion channels and transporters in cell migration. Physiol Rev. 2012;92:1865‐1913. [DOI] [PubMed] [Google Scholar]
- 30. de Lucas B, Perez LM, Galvez BG. Importance and regulation of adult stem cell migration. J Cell Mol Med. 2018;22:746‐754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Mayor R, Etienne‐Manneville S. The front and rear of collective cell migration. Nat Rev Mol Cell Biol. 2016;17:97‐109. [DOI] [PubMed] [Google Scholar]
- 32. Jiang L‐H, Mousawi F, Yang X, Roger S. ATP‐induced Ca2+‐signalling mechanisms in the regulation of mesenchymal stem cell migration. Cell Mol Life Sci. 2017;74:3697‐3710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Burnstock G, Ulrich H. Purinergic signaling in embryonic and stem cell development. Cell Mol Life Sci. 2011;68:1369‐1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Jiang L‐H, Hao Y, Mousawi F, Peng H, Yang X. Expression of P2 purinergic receptors in mesenchymal stem cells and their roles in extracellular nucleotide regulation of cell functions. J Cell Physiol. 2017;232:287‐297. [DOI] [PubMed] [Google Scholar]
- 35. Riddle RC, Taylor AF, Rogers JR, Donahue HJ. ATP release mediates fluid flow‐induced proliferation of human bone marrow stromal cells. J Bone Miner Res. 2007;22:589‐600. [DOI] [PubMed] [Google Scholar]
- 36. Coppi E, Pugliese AM, Urbani S, et al. ATP modulates cell proliferation and elicits two different electrophysiological responses in human mesenchymal stem cells. Stem Cells. 2007;25:1840‐1849. [DOI] [PubMed] [Google Scholar]
- 37. Zippel N, Limbach CA, Ratajski N, et al. Purinergic receptors influence the differentiation of human mesenchymal stem cells. Stem Cells Dev. 2012;21:884‐900. [DOI] [PubMed] [Google Scholar]
- 38. Sun D, Junger WG, Yuan C, et al. Shockwaves induce osteogenic differentiation of human mesenchymal stem cells through ATP release and activation of P2X7 receptors. Stem Cells. 2013;31:1170‐1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ferrari D, Gulinelli S, Salvestrini V, et al. Purinergic stimulation of human mesenchymal stem cells potentiates their chemotactic response to CXCL12 and increases the homing capacity and production of proinflammatory cytokines. Exp Hematol. 2011;39:360‐374, 374.e361‐365. [DOI] [PubMed] [Google Scholar]
- 40. Peng H, Hao Y, Mousawi F, et al. Purinergic and store‐operated Ca2+ signaling mechanisms in mesenchymal stem cells and their roles in ATP‐induced stimulation of cell migration. Stem Cells. 2016;34:2102‐2114. [DOI] [PubMed] [Google Scholar]
- 41. Goetzke R, Sechi A, De Laporte L, et al. Why the impact of mechanical stimuli on stem cells remains a challenge. Cell Mol Life Sci. 2018;75:3297‐3312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Liu YS, Lee OK. In search of the pivot point of mechanotransduction: mechanosensing of stem cells. Cell Transplant. 2014;23:1‐11. [DOI] [PubMed] [Google Scholar]
- 43. Coste B, Mathur J, Schmidt M, et al. Piezo1 and Piezo2 are essential components of distinct mechanically activated cation channels. Science. 2010;330:55‐60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Li J, Hou B, Tumova S, et al. Piezo1 integration of vascular architecture with physiological force. Nature. 2014;515:279‐282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Pathak MM, Nourse JL, Tran T, et al. Stretch‐activated ion channel Piezo1 directs lineage choice in human neural stem cells. Proc Natl Acad Sci USA. 2014;111:16148‐16153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Lukacs V, Mathur J, Mao R, et al. Impaired PIEZO1 function in patients with a novel autosomal recessive congenital lymphatic dysplasia. Nat Commun. 2015;6:8329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Cahalan SM, Lukacs V, Ranade SS, Chien S, Bandell M, Patapoutian A. Piezo1 links mechanical forces to red blood cell volume. Elife. 2015;4:e07370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Wang S, Chennupati R, Kaur H, Iring A, Wettschureck N, Offermanns S. Endothelial cation channel PIEZO1 controls blood pressure by mediating flow‐induced ATP release. J Clin Investig. 2016;126:4527‐4536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Wang Y, Chi S, Guo H, et al. A lever‐like transduction pathway for long‐distance chemical‐ and mechano‐gating of the mechanosensitive Piezo1 channel. Nat Commun. 2018;9:1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Zhang J, Zhou Y, Huang T, et al. PIEZO1 functions as a potential oncogene by promoting cell proliferation and migration in gastric carcinogenesis. Mol Carcinog. 2018;57:1144‐1155. [DOI] [PubMed] [Google Scholar]
- 51. Zhao Q, Zhou H, Chi S, et al. Structure and mechanogating mechanism of the Piezo1 channel. Nature. 2018;554:487‐492. [DOI] [PubMed] [Google Scholar]
- 52. Wu J, Lewis AH, Grandl J. Touch, tension, and transduction ‐ the function and regulation of Piezo ion channels. Trends Biochem Sci. 2017;42:57‐71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Saotome K, Murthy SE, Kefauver JM, Whitwam T, Patapoutian A, Ward AB. Structure of the mechanically activated ion channel Piezo1. Nature. 2018;554:481‐486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Miyamoto T, Mochizuki T, Nakagomi H, et al. Functional role for Piezo1 in stretch‐evoked Ca2+ influx and ATP release in urothelial cell cultures. J Biol Chem. 2014;289:16565‐16575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Cinar E, Zhou S, DeCourcey J, Wang Y, Waugh RE, Wan J. Piezo1 regulates mechanotransductive release of ATP from human RBCs. Proc Natl Acad Sci USA. 2015;112:11783‐11788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Zhang T, Chi S, Jiang F, Zhao Q, Xiao B. A protein interaction mechanism for suppressing the mechanosensitive Piezo channels. Nat Commun. 2017;8:1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Li C, Rezania S, Kammerer S, et al. Piezo1 forms mechanosensitive ion channels in the human MCF‐7 breast cancer cell line. Sci Rep. 2015;5:8364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Yang XN, Lu YP, Liu JJ, et al. Piezo1 is as a novel trefoil factor family 1 binding protein that promotes gastric cancer cell mobility in vitro. Dig Dis Sci. 2014;59:1428‐1435. [DOI] [PubMed] [Google Scholar]
- 59. McHugh BJ, Murdoch A, Haslett C, et al. Loss of the integrin‐activating transmembrane protein Fam38A (Piezo1) promotes a switch to a reduced integrin‐dependent mode of cell migration. PLoS One. 2012;7:e40346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Sugimoto A, Miyazaki A, Kawarabayashi K, et al. Piezo type mechanosensitive ion channel component 1 functions as a regulator of the cell fate determination of mesenchymal stem cells. Sci Rep. 2017;7:17696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Gao Q, Cooper PR, Walmsley AD, Scheven BA. Role of Piezo channels in ultrasound‐stimulated dental stem cells. J Endod. 2017;43:1130‐1136. [DOI] [PubMed] [Google Scholar]
- 62. Tang JM, Yuan J, Li Q, et al. Acetylcholine induces mesenchymal stem cell migration via Ca2+/PKC/ERK1/2 signal pathway. J Cell Biochem. 2012;113:2704‐2713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Lin CY, Zu CH, Yang CC, et al. IL‐1beta‐induced mesenchymal stem cell migration involves MLCK activation via PKC signaling. Cell Transplant. 2015;24:2011‐2028. [DOI] [PubMed] [Google Scholar]
- 64. Yamamoto S, Shimizu S, Kiyonaka S, et al. TRPM2‐mediated Ca2+ influx induces chemokine production in monocytes that aggravates inflammatory neutrophil infiltration. Nat Med. 2008;14:738‐747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Lev S, Moreno H, Martinez R, et al. Protein tyrosine kinase PYK2 involved in Ca2+‐induced regulation of ion channel and MAP kinase functions. Nature. 1995;376:737‐745. [DOI] [PubMed] [Google Scholar]
- 66. Syed Mortadza SA, Sim JA, Stacey M, et al. Signalling mechanisms mediating Zn2+‐induced TRPM2 channel activation and cell death in microglial cells. Sci Rep. 2017;7:45032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Syed Mortadza SA, Sim JA, Neubrand VE, et al. A critical role of TRPM2 channel in Aβ42‐induced microglial activation and generation of tumor necrosis factor‐α. Glia. 2018;66:562‐575. [DOI] [PubMed] [Google Scholar]
- 68. Jiang L‐H, Li X, Syed Mortadza SA, Lovatt M, Yang W. The TRPM2 channel nexus from oxidative damage to Alzheimer's pathologies: an emerging novel intervention target for age‐related dementia. Ageing Res Rev. 2018;47:67‐79. [DOI] [PubMed] [Google Scholar]
- 69. Li X, Jiang L‐H. Multiple molecular mechanisms form a positive feedback loop driving amyloid beta42 peptide‐induced neurotoxicity via activation of the TRPM2 channel in hippocampal neurons. Cell Death Dis. 2018;9:195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Chen YF, Chiu WT, Chen YT, et al. Calcium store sensor stromal‐interaction molecule 1‐dependent signaling plays an important role in cervical cancer growth, migration, and angiogenesis. Proc Natl Acad Sci USA. 2011;108:15225‐15230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Heid CA, Stevens J, Livak KJ, Williams PM. Real time quantitative PCR. Genome Res. 1996;6:986‐994. [DOI] [PubMed] [Google Scholar]
- 72. Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real‐time quantitative PCR and the 2‐ΔΔCt method. Methods. 2001;25:402‐408. [DOI] [PubMed] [Google Scholar]
- 73. Syeda R, Xu J, Dubin AE, et al. Chemical activation of the mechanotransduction channel Piezo1. Elife. 2015;4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Bae C, Sachs F, Gottlieb PA. The mechanosensitive ion channel Piezo1 is inhibited by the peptide GsMTx4. Biochemistry. 2011;50:6295‐6300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Jensen ME, Odgaard E, Christensen MH, et al. Flow‐induced [Ca2+]i increase depends on nucleotide release and subsequent purinergic signaling in the intact nephron. J Am Soc Nephrol. 2007;18:2062‐2070. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure 1. RT‐qPCR analysis of Piezo1 mRNA expression in human DP‐MSCs.
Representative RT‐qPCR amplification plots (left) and mean Piezo1/β‐actin (right) from 3 independent mRNA preparations for 9F, and 2 independent mRNA preparations for 22M, 20M and 32F cells.
Figure 2. Effects of ruthenium red on Yoda1‐induced Ca 2+ responses in human DP‐MSCs.
Representative Yoda1‐induced intracellular Ca2+ responses to 3 μM Yoda1 in cells from 20M (A) or 32F (B) without (CTL) or after treatment with 10 or 30 μM ruthenium red (RR) for 5 minutes from one set of experiments using 4 wells of cells for each condition.
Figure 3. Effect of GsMTx4 on Yoda1‐induced Ca 2+ responses in human DP‐MSCs.
Representative Yoda1‐induced intracellular Ca2+ responses to 3 μM Yoda1 in cells from 20M without (CTL) and with treatment with 1 or 3 μM GsMTx4 for 30 minutes prior and during exposure to Yoda1 from one set of experiments using 4 wells of cells for each condition.
Figure 4. Effect of siRNA‐mediated knockdown of Piezo1 expression on Yoda1‐induced Ca 2+ response in hDP‐MSCs
Representative intracellular Ca2+ responses to 3 μM Yoda1 from one set of experiments using 4 wells of cells for each condition, in cells from 9F that were transfected with control siRNA (siCTL) or Piezo1‐specific siRNA (siPiezo1). Cells were exposed to 5 μM ionomycin at the end of recordings.
Figure 5. No effect of apyrase or PPADS on human DP‐MSC migration.
Summary of mean wound narrowing in 3 independent experiments for cells from 9F under control condition (CTL) and cells exposed to 0.3 or 1 U/mL apyrase (Apy) (A) or 30 μM PPADS in cells from 9F and 22M (B). NS, no significant difference.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.


