FIG 1.
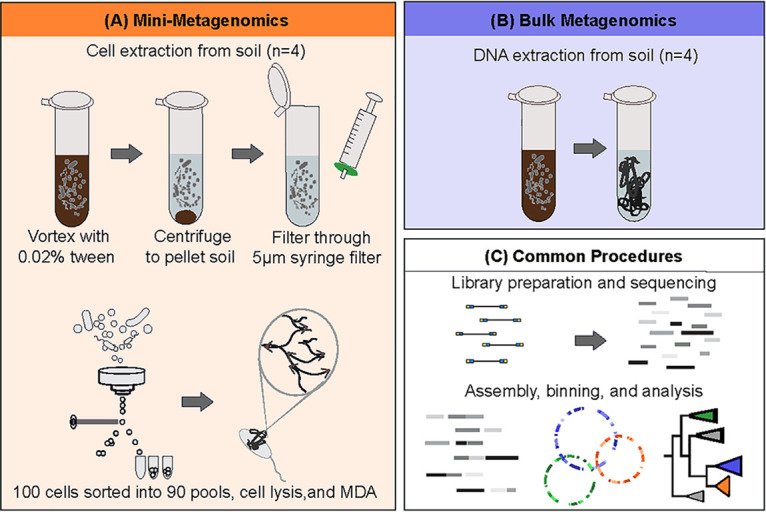
Overview of mini-metagenome and bulk metagenome approaches used in this study. (A) Mini-metagenomics performed on four soil samples, including one heated sample from the top organic soil, one heated sample from the lower mineral soil, one control organic sample, and one control mineral sample (n = 4). Cells were separated from soil particles using a mild detergent, followed by vortex mixing, centrifugation, and filtration through a 5-μm-pore-size syringe filter. Suspended cells were stained with SYBR green and sorted into 90 pools of 100 cells each, generating 359 mini-metagenomes. (B) Bulk metagenomic sequencing conducted on the four soils that were used in mini-metagenomics. (C) Following nucleic acid extraction, libraries were prepared, and shotgun sequencing was performed. Sequence data underwent assembly and quality control. Data were binned and assessed for bin quality. Only medium-quality genome bins with estimates of 50% completeness, 10% contamination, and 10% strain heterogeneity were used in downstream phylogenomic and functional analyses. Further details are provided in Materials and Methods.
