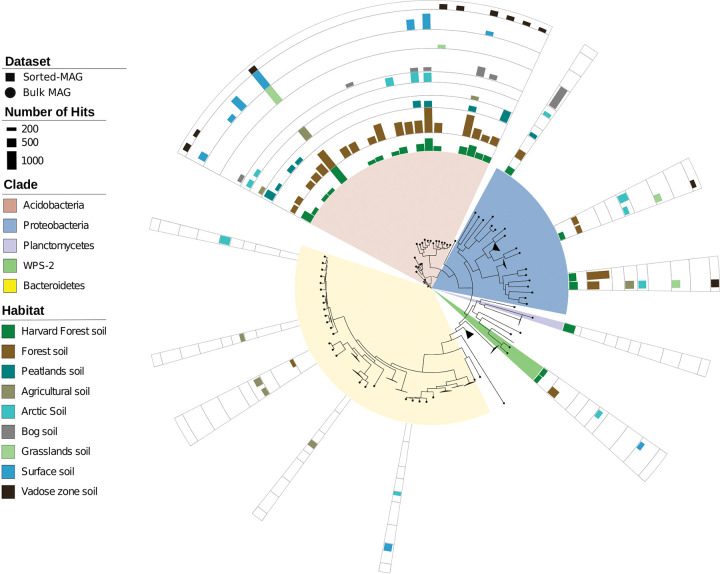
FIG 4.
Comparison of MAGs from this study with published data from terrestrial metagenomes. Innermost is a maximum likelihood tree based on a concatenated alignment of 56 conserved marker proteins from medium-quality sorted-MAGs and bulk MAGs recovered in this study. Mini-metagenomes and bulk MAGs were dereplicated by clustering at 95% average nucleotide identity, resulting in 173 sorted-MAGs and 28 bulk MAGs. The clade names are color-coded according to phylum. Individual tracks around the tree depict hits of individual sorted-MAGs and bulk MAGs by metagenome samples arising from each terrestrial habitat type as specified in the legend. The height of the bar chart indicates the total number of sorted-MAGs and bulk MAG coding sequences that matched metagenome samples. The MAGs were considered matches if they had a minimum of 200 coding sequences with hits at ≥95% amino acid identity over 70% alignment lengths to CDS of an individual metagenome. Further details are provided in Materials and Methods, and data corresponding to this figure are provided in Table S3. The figure was rendered using iTOL (96).