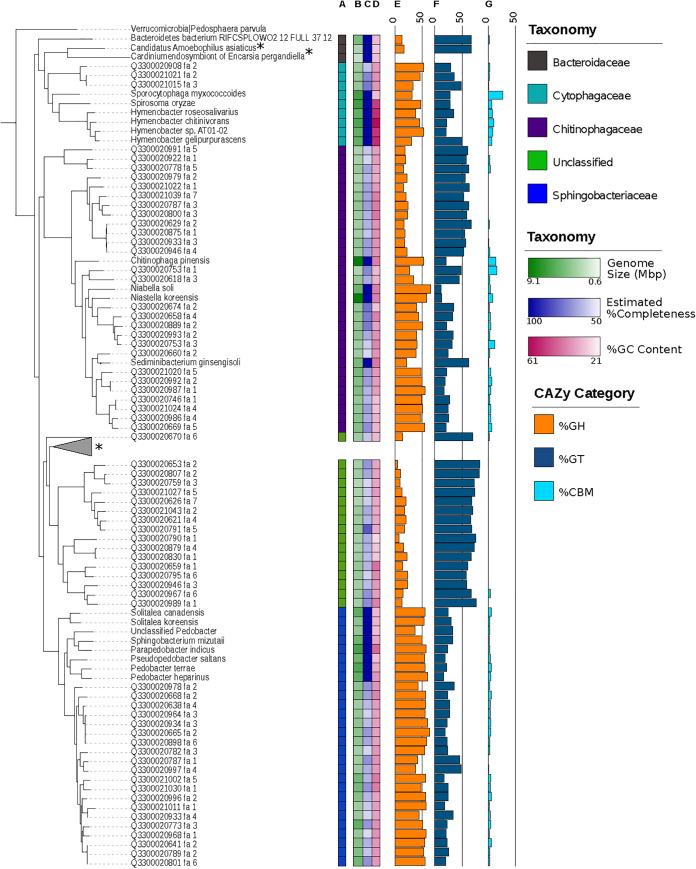
FIG 5.
Insights into carbon metabolism within the phylum Bacteroidetes. A concatenated marker gene tree of 67 Bacteroidetes sorted-MAGs and 70 Bacteroidetes reference sequences from the IMG/M database shows clade-specific abundances of glycoside hydrolase and glycosyl transferases. The tree is rooted with Pedosphaera parvula (phylum Verrucomicrobia). Column A shows the distribution of sorted-MAGs across three families of Bacteroidetes, including Cytophagaceae, Chitinophagaceae, and Sphingobacteriaceae, and a clade of unclassified sorted-MAGs. Column B shows genome sizes, with the darkest color representing the largest genome of 9.1 megabases and the lightest representing a genome size of 0.6 megabases. Column C shows genome completeness based on CheckM marker genes, ranging from 50% to 80.5%, as a color gradient. Reference sequences represent isolates with complete genomes. Column D presents genome GC content as a color gradient that ranges from 21.13% to 61.24%. In columns E to G, percentages of genes annotated as glycoside hydrolases (column E), glycosyl transferases (column F), and carbohydrate binding modules (column G) are illustrated as bar charts with vertical lines denoting 0% and 50% of annotated genes. Bacteroidetes with known symbiotic relationships are indicated with an asterisk. The collapsed clade contains Sulcia muelleri, a known symbiont of sap-feeding insects, and Blatellabacterium sp., a known symbiont of the cockroach Blatella germanica.