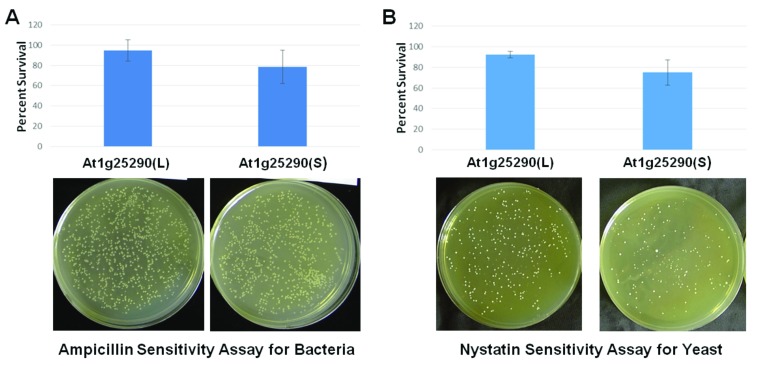
Figure 10. Activity assays using At1g25290 splice variant proteins as exogenous additives to bacteria and yeast cells.
( A) Recombinant At1g25290 splice variant proteins L and S were tested for activity (for enhanced sensitivity to ampicillin in this case) as exogenous additives and bacteria. The assays were conducted in the same manner as that shown in Figure 8 (from untreated to mock to added proteins). Cells (resulting colonies) surviving the different treatments were then assessed and represented as Percent Survival. The key results comparing the splice variants L and S are shown in this panel. The bar graphs are arranged to the corresponding representative results, the resulting agar plates. The error bars represent standard deviations (n=4, T-test, P=0.01). ( B) Recombinant At1g25290 splice variant proteins L and S were also tested for activity (for enhanced sensitivity to nystatin in this case) as exogenous additives and yeast. The assays were conducted in the same manner as that shown in Figure 9 (from untreated to mock to added proteins). The organization of panel B is the same as in panel A, except for yeast and nystatin. The error bars represent standard deviations (n=4, T-test, P=0.01).