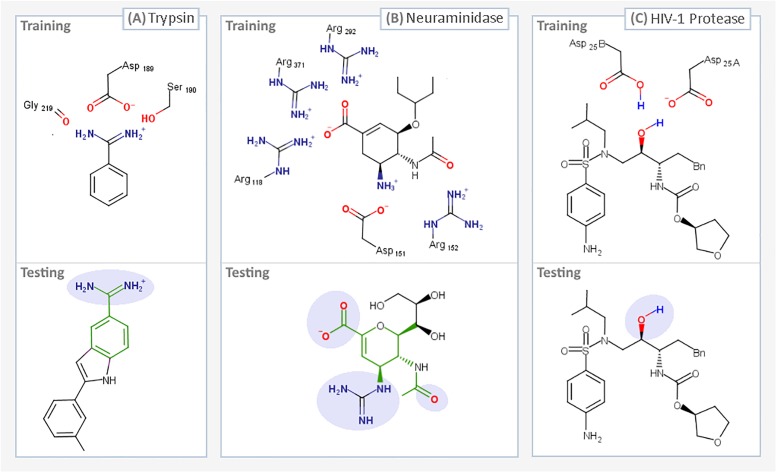
Fig 2. Summary of iMD-VR docking tasks.
The ligands that were interactively unbound and rebound in a series of user tests, two for each of the three systems: Trypsin (A), Neuraminidase (B), and HIV-1 Protease (C). For each protein system, we devised two docking tasks, a training phase (where a trace representation of the ligand in the bound pose was present to guide users as shown in supplementary videos A–C in S1 File), and a testing phase, where no trace atoms were shown. For the testing tasks, parts of the ligand which interact with key residues have been highlighted in lilac. For proteins A and B, where the testing task ligand differs from training, the shared scaffold between the two ligands is highlighted in green.