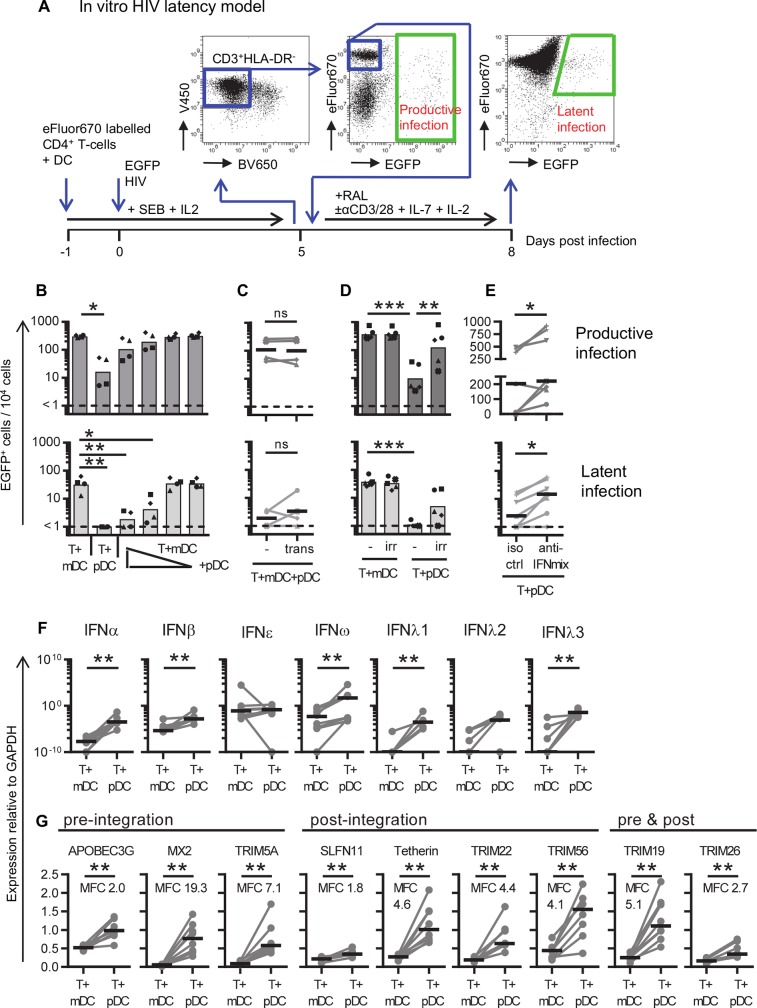
Fig 1. pDC-induced inhibition of HIV latency.
A: Schematic of the in vitro DC-T cell HIV latency model. Resting CD4+ T cells were negatively selected using magnetic cell sorting, stained with the proliferation dye eFluor670 and cultured with and without syngeneic sorted DC subsets (DC:T cell ratio of 1:10) for 24 hrs. in the presence of staphylococcal enterotoxin B (SEB) and IL-2. Cells were infected with CCR5-using full-length nef-competent EGFP-reporter virus (EGFP HIV). At day 5 post-infection, EGFP+ cells were measured by flow cytometry and used as a measure for productive infection. The CD3+HLA-DR- non-productively infected (EGFP-), non-proliferating (eFluor670HI) CD4+ T cells were sorted and cultured with an HIV integrase inhibitor (raltegravir; RAL) in the presence or absence of activation stimuli (anti-CD3/CD28+IL-7+IL-2) for 72 hrs. After stimulation, EGFP expression was measured by flow cytometry and latent infection quantified as the number of EGFP+ cells in the stimulated culture after subtracting the number of EGFP+ cells in the unstimulated culture (background). Resting CD4+ T cells were co-cultured with B: mDC, pDC, or both at different ratios of mDC:pDC (1:1, 10:1, 100:1 and 1,000:1, n = 4), C: mDC and pDC were co-cultured at a 1:1 ratio, in the absence (-) or presence of a transwell (trans, n = 4), D: T cells were co-cultured with mDC or pDC that were irradiated (irr) or not (-, n = 5, DC:T cell ratio of 1:10), E: T cells were cultured with pDC in the presence of antibodies blocking IFNAR and neutralizing soluble IFNα and IFNβ (anti-IFNmix) or an isotype control (iso ctrl, n = 7, DC:T cell ratio of 1:10). Productive (dark grey, top panels) and latent (light grey, bottom panels) infection in non-proliferating cells was quantified and shown in panels B-E. F,G: Resting CD4+ T cells were co-cultured with pDC or mDC (DC:T cell ratio of 1:10), infected with EGFP HIV, harvested at 1 day post-infection, RNA was extracted and reverse transcribed into cDNA for qPCR detection of F: type I IFNs (IFNα, IFNβ, IFNε, IFNω) and type III IFNs (IFNλ1/IL-29, IFNλ2/IL-28A, IFNλ3/IL-28B, n = 8) or G: virus restriction factors that inhibit HIV replication prior to integration, post-integration, or both (n = 8, MFC = mean fold change). Values were normalized to mRNA of the house keeping gene GAPDH. Columns or lines represent mean values (n≤5) or median values (n>5). Symbols and dots represent individual donors. *p<0.05, **p<0.01, ***p<0.001, ns = not significant, as determined by paired student T test (n≤5) on log-transformed data or Wilcoxon matched pairs signed rank test (n>5).