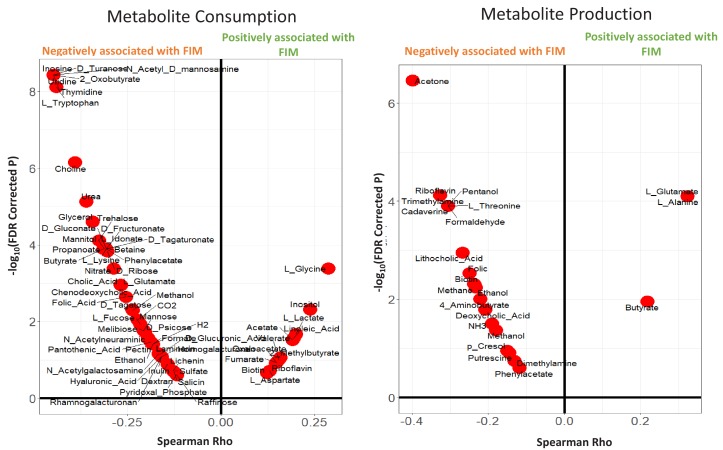
Figure 6. Frailty-associated markers have shared positive associations across multiple diseases in both age groups and have a specific metabolic signature.
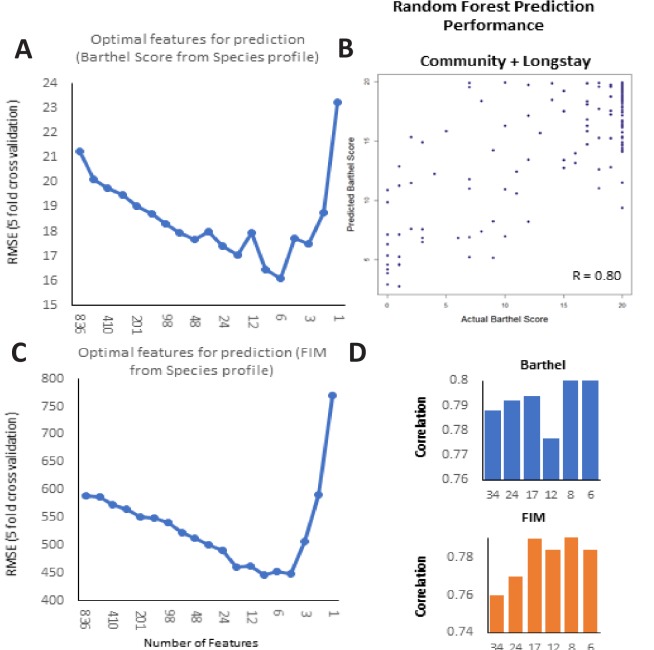
(A) Actual FIM values versus FIMs predicted by Random forest of microbiome features of the elderly individuals of the ELDERMET cohort living in Community or Residential care (Longstay). (B) Mean ranks of the various taxonomic groups (identified in Figure 3) for the prediction of FIM (an inverse measure of frailty) in the ELDERMET cohort. (C) Variable Importance Scores of the eight markers with the highest predictive power in the Random Forest models for prediction of FIM. A comparison of the abundance of markers between HighFIM and LowFIM individuals indicated that all of these markers were associated with frailty state. (D) The network in the central panel indicates the 13 metabolite profiles significantly associated with the top markers. Taxon markers are indicated in the center. Consumption profiles are in the upper half (in pink octagons) and Production profiles are on the lower panel (in yellow octagons). Edges indicate presence. Second from the left in top panel are the correlations between predicted and actual FIM values obtained for iterative bootstrapped Random Forest models (training on 20% and testing on the rest 80%) using only the 13 metabolite profile markers of (D), all metabolite profiles and all metabolite profiles removing the 13 metabolite markers. Top and bottom panels show the validation (indicated by arrows) obtained for the predicted metabolite markers using either the measured metabolites, dietary consumption profiles, specific microbial pathway abundances as well as the CutC gene family abundances identified using humann2 (shown either as boxplot comparing the profiles between Frail and Non-Frail individuals or scatterplots showing correlations between the measured metabolite level and the FIM value of the individuals). A total of 11 of the 13 metabolites could be validated using either of these strategies.