Fig. 2. Evolutionary comparison and gene conservation of the jojoba (S. chinensis) genome.
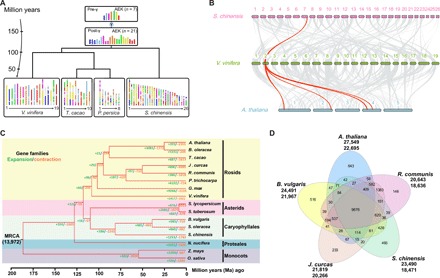
(A) Evolutionary scenario of S. chinensis from the AEK of 7 (pre-γ) and 21 (post-γ) protochromosomes reconstructed from a comparison of the V. vinifera (grape), T. cacao (chocolate), and P. persica (peach) genomes. The modern genomes (bottom) are illustrated with different colors reflecting the seven ancestral chromosomes of AEK origin (top). γ refers to the whole-genome triplication (γ) shared among the eudicots. (B) Macrosynteny between genomic regions of S. chinensis, V. vinifera, and A. thaliana. Macrosynteny patterns between jojoba and grape show that each jojoba region aligns with one region in grape, and each grape region aligns to four syntenic regions in A. thaliana that experienced two additional rounds of crucifer genome duplication. (C) Expansion and contraction of gene families among 16 plant species. The number at the root (13,972) denotes the total number of gene families predicted in the most recent common ancestor (MRCA). A total of 1253 gene families are substantially expanded, and 1783 gene families are contracted in S. chinensis compared with other plant genomes. (D) Shared gene families among S. chinensis, A. thaliana, B. vulgaris, J. curcas, and R. communis. The five species contain 9876 common gene families, and S. chinensis has 493 specific gene families.
