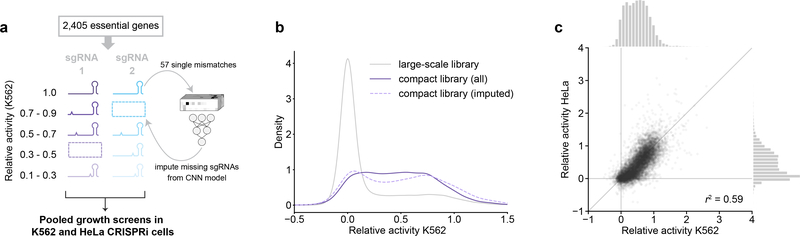
Figure 5.
Compact mismatched sgRNA library targeting essential genes. (a) Design of library. For activity bins lacking a previously measured sgRNA, novel mismatched sgRNAs were included according to predicted activity. (b) Distribution of relative activities from the large-scale library (gray) and the compact library (purple) in K562 cells. The dashed line represents sgRNAs that were selected based on predicted activity from the deep learning model. (c) Comparison of relative activities of mismatched sgRNAs in HeLa and K562 cells. Marginal histograms show the distributions of relative activities along the corresponding axes. n = 9,514 sgRNAs; r2 = squared Pearson correlation coefficient.