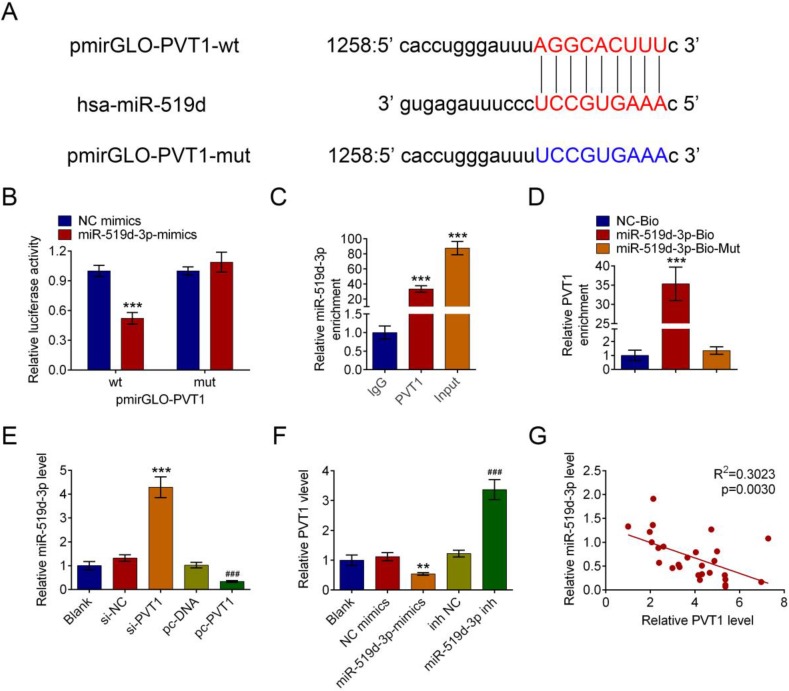
Figure 4.
PVT1 negatively regulates miR-159-3p in PDAC cells. (A) The putative binding site of miR-159-3p and PVT1 was predicted by microrna.org. (B) The luciferase activity of PVT1 was detected using the luciferase report gene assay in HPAC cells co-transfected the wild type (pmirGLO-PVT1-wt) or mutated reporter construct (pmirGLO-PVT1-mut) and miR-159-3p-mimics or NC mimics. (C) Cellular lysates from HPAC cells were used for RIP with an Ago2 antibody and IgG antibody. The levels of miR-159-3p were detected by qRT-PCR. (D) HPAC cells were transfected with biotinylated NC (NC-Bio), biotinylated wild-type miR-519d-3p (miR-519d-3p-Bio) or biotinylated mutant miR-138 (miR-519d-3p-Bio-Mut), and biotin-based miRNA pull-down assays were conducted after 48 h of transfection. The PVT1 were analysed by qRT-PCR. (E) The miR-159-3p were detected in PDAC cells transfected with si-NC, si-PVT1#3, pc-DNA3.1 or pc-DNA3.1-PVT1. The PVT1 were detected in PDAC cells transfected with NC-mimics, miR-159-3p-mimics, inh NC or miR-159-3p-inh. (F) Correlation between PVT1 and miR-159-3p were detected in the cancer tissue of PDAC patients. (** and ## P < 0.01, ***and ### P < 0.001)