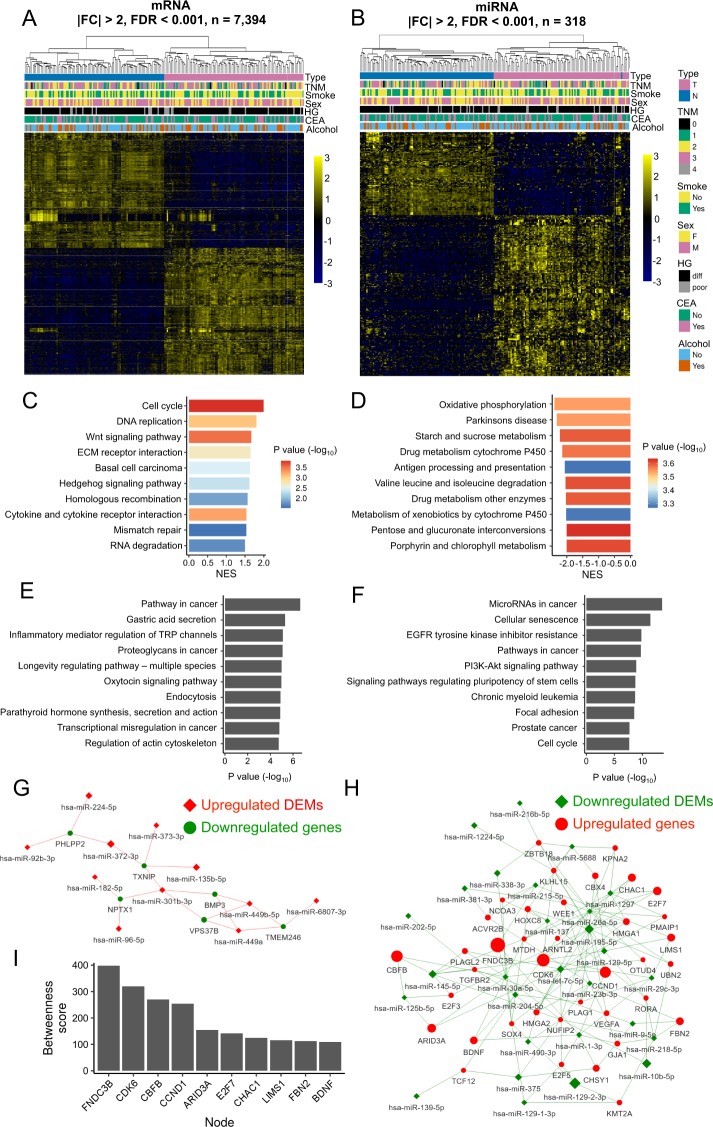
Figure 1.
Comprehensive profiling of Taiwanese CRC transcriptomes reveals cohort-specific alterations. Hierarchical clustering of (A) DEGs and (B) DEMs from Taiwanese CRC patients demonstrating a disturbed expression profile between normal (blue) and tumor (red) tissues. GSEA charts of top ten significantly enriched (P value < 0.05) upregulated (C, NES > 0) and downregulated (D, NES < 0) KEGG pathways, where each row represents indicated NES value and the color shows the significance of enrichment result in -log10 transformed P value. Detailed enrichment results are listed in the Table S3, Additional file 2. Gene set over-representation analysis of top ten (E) upregulated and (F) downregulated DEMs targeting mRNAs enriched pathways. Construction of miRNA–mRNA correlation network based on (G) upregulated DEMs/downregulated DEGs and (H) downregulated DEMs/upregulated DEGs, in which DEGs are targeted by more than two miRNAs. miRNAs, squares; mRNA, circles; upregulated, red; downregulated, green. (I) Bar plot of betweenness scores for top ten nodes from networks (G) and (H).