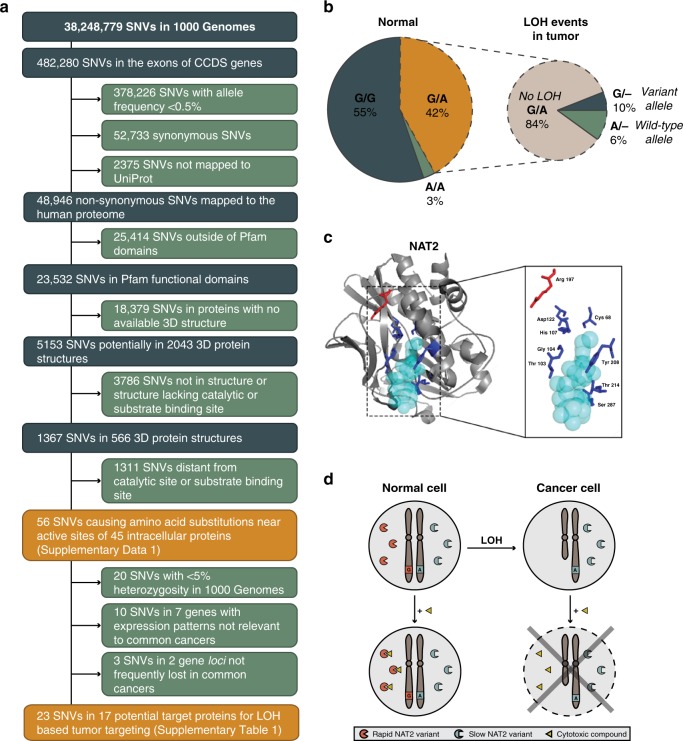
Fig. 1. N-acetyltransferase 2 (NAT2) is a target for allele-specific inhibition.
a Identification of enzymes with prevalent alternative alleles resulting in amino-acid substitution near functional sites. A series of filtering steps was applied to identify the subset of SNVs causing amino-acid substitutions near active sites in publicly available 3D protein structures. To enrich potential target proteins for LOH-based tumor targeting approaches, the candidate genes were further selected based on expression profile and the frequency of LOH in common cancer types. b Loss of heterozygosity at 8p22 can render CRCs deficient in NAT2 function. To confirm the prevalence of rs1799930 and the frequency of LOH at NAT2, genomic DNA from 74 CIN CRCs and patient-matched normal tissues was genotyped. Left, genotype distribution of normal tissues. Right, LOH events observed in the tumors of heterozygous individuals. c Structure of human NAT2 co-crystallized with the cofactor coenzyme A (light blue) (PDB: 2PFR). The side chains of the active site amino-acid residues involved in substrate transformation (Asp122, Cys68, and His107) and cofactor binding (Gly104, Thr103, Thr214, Tyr208, and Ser287) (dark blue) and the side chain of Arg197 (rs1799930, red) are shown. The figure was designed in PyMOL (v. 2.3.2). d Exploiting loss of a rapid NAT2 allele in tumor cells for anti-cancer therapy. Eligible patients are heterozygous for the slow (A, blue) and rapid (G, red) NAT2 alleles. During cancer progression, cancer cells may undergo loss of heterozygosity (LOH) and lose the rapid NAT2 allele (red). Treatment with a cytotoxic compound (triangle) that can only be processed by the rapid NAT2 enzymatic variant lost in cancer cells will result in selective tumor death.