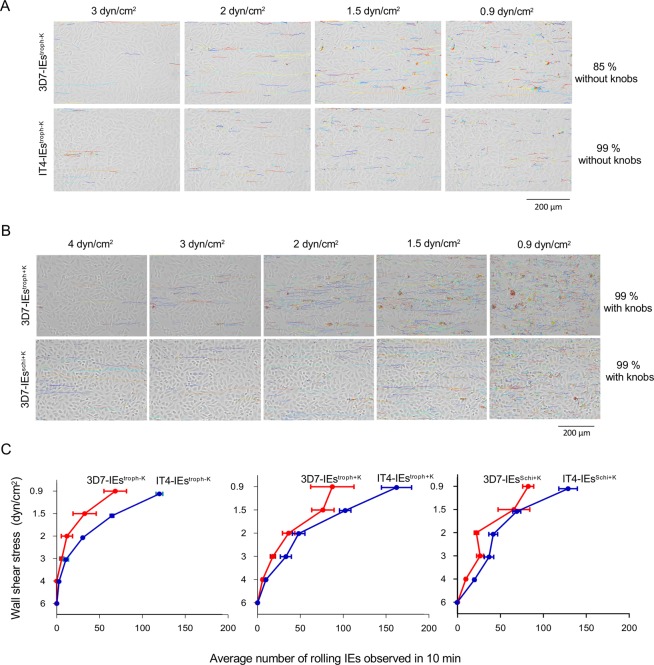
Figure 2.
Cell trajectories showing the rolling binding behavior of IEs to CHO-745CD36. Trajectories of 3D7-IEstroph−K and IT4-IEstroph−K. (A) Trajectories of 3D7-IEstroph+K and 3D7-IEsschi+K under various shear stresses. (B) Average number of rolling IEs at applied shear stresses. (C) Images were analyzed using the trackdem (0.4.2) R package38,58. The static cell background was subtracted from all images and then moving particles (i.e., IEs) were detected. The Shiny R package38,58 was used with thresholding of 0.1. The PixelRange of moving particles was set to 30–500. Individual trajectories were subsequently reconstructed based on a minimum present (incThres) equal to six frames (i.e., 30 sec). Finally, trajectories were plotted over the still background. Examples of trajectories are shown in S1 and S2 Movies.