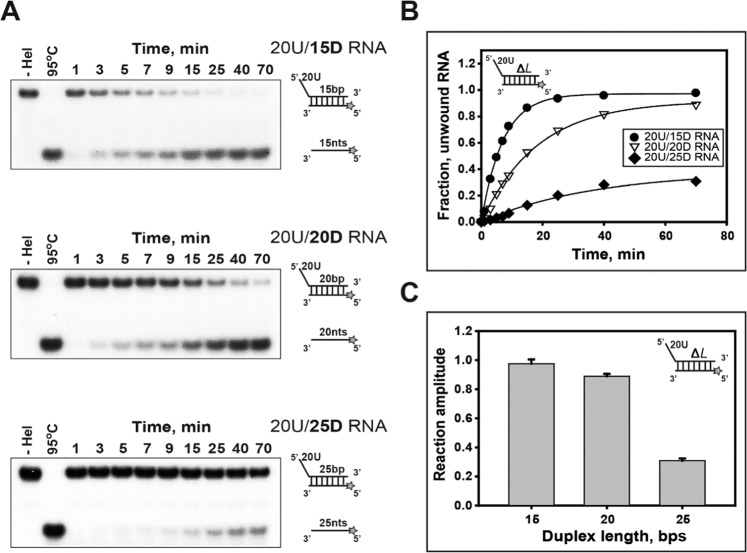
Figure 2.
Processivity on duplex RNA substrates with different duplex lengths. (A,B): The single-turnover unwinding of duplex RNA substrates with different lengths of duplex. (A) Gel retardation assay of nsP13 and duplex RNAs with different duplex lengths. The substrates of 15 bp (15D), 20 bp (20D), and 25 bp (25D) duplexes containing a 5′-poly(U) tail of 20 bases (20U) were designed as shown in Supplementary Table 1. The unwinding products were resolved by non-denaturing 15% PAGE. (B, C) The amplitudes were as follows: 15 bp (●: 20U/15D RNA) = 0.98 ± 0.03, 20 bp (▽: 20U/20D RNA) = 0.89 ± 0.02, and 25 bp (◆: 20U/25D RNA) = 0.31 ± 0.02.