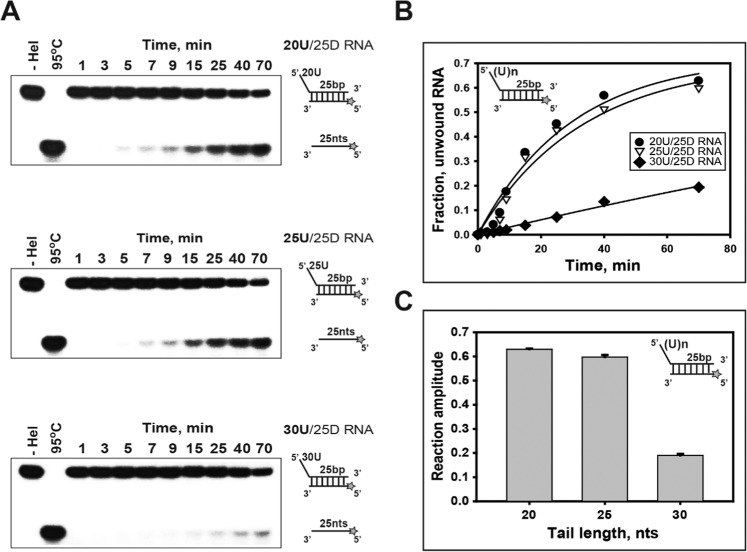
Figure 3.
Unwinding of duplex RNA substrates with 5′-ss tails of different lengths. (A,B): Single-turnover unwinding of 25 bp (25D) RNA substrates with 5′-poly(U) tails of different lengths by helicase nsP13. (A) Gel retardation assay of nsP13 and duplex RNA substrates with 5′-ss tails of different lengths. Three different substrates containing 25 duplex-fixed (25D) and the 5′-poly(U) tail of 20 bases (20U), 25 bases (25U), and 30 bases (30U) were designed as shown in Supplementary Table 1. The unwinding products were resolved by non-denaturing 15% PAGE. (B,C) The amplitudes were as follows: 20U (●: 20U/25D RNA) = 0.63 ± 0.01, 25U (▽: 25U/25D RNA) = 0.6 ± 0.01, and 30U (◆: 30U/25D RNA) = 0.19 ± 0.01.