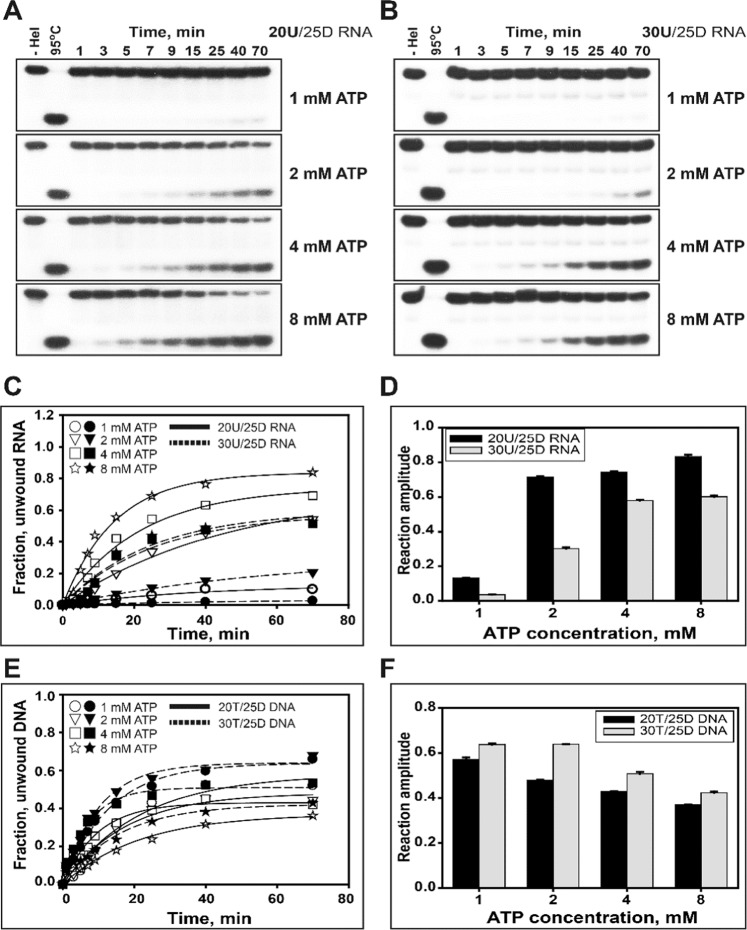
Figure 6.
Additional ATP requirement to promote ATP-dependent translocation of helicase nsP13. (A,B): Gel retardation assay of helicase nsP13 and duplex RNA substrates. The unwinding of 20U/25D RNA and 30U/25D RNA or 20 T/25D DNA and 30 T/25D DNA was reacted with various concentrations of ATP (1, 2, 4, and 8 mM). The unwinding reaction was performed with 0.5 μM nsP13 (for RNA) or 0.1 μM nsP13 (for DNA) and each 5 nM substrate at 37 °C as described in the standard unwinding method. The unwinding products were resolved by non-denaturing 15% PAGE. (C,D) The amplitudes for RNAs were as follows: ①20U/25D RNA (Solid line) – 1 mM ATP (○) = 0.13 ± 0.001, 2 mM ATP (▽) = 0.71 ± 0.005, 4 mM ATP (□) = 0.74 ± 0.005, and 8 mM ATP (☆) = 0.83 ± 0.01; ②30U/25D RNA (Dotted line) – 1 mM ATP (●) = 0.03 ± 0.001, 2 mM ATP (▼) = 0.3 ± 0.01, 4 mM ATP(■) = 0.58 ± 0.003, and 8 mM ATP (★) = 0.6 ± 0.01. (E, F) The amplitudes for DNAs were as follows: ①20 T/25D DNA (Solid line) – 1 mM ATP (○) = 0.57 ± 0.01, 2 mM ATP (▽) = 0.48 ± 0.001, 4 mM ATP (□) = 0.43 ± 0.001, and 8 mM ATP (☆) = 0.37 ± 0.002; ②30 T/25D DNA (Dotted line) – 1 mM ATP (●) = 0.64 ± 0.004, 2 mM ATP (▼) = 0.64 ± 0.001, 4 mM ATP (■) = 0.51 ± 0.01, and 8 mM ATP (★) = 0.42 ± 0.01.