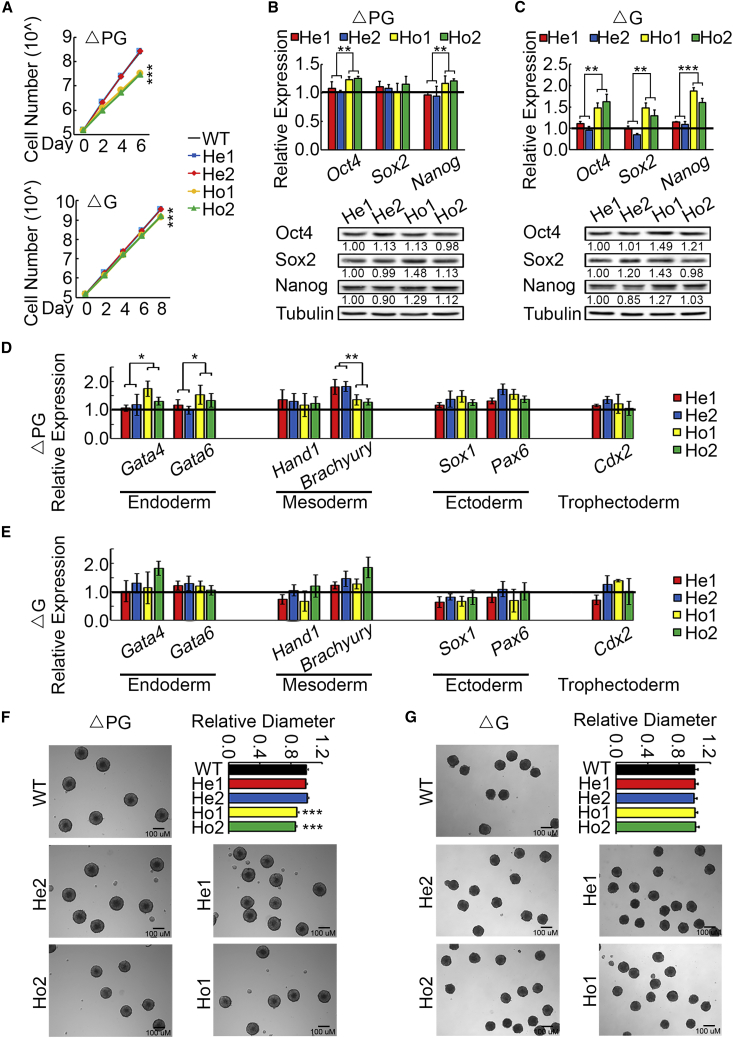
Figure 3.
Homozygous ΔPG ESCs Display a More Severe Proliferation Defect than Homozygous ΔG ESCs
(A) Proliferation assays for ΔPG, ΔG, and WT ESCs. The Y axis is log scale of cell number. Three biological replicates per sample were assayed. Data are shown as the mean ± SEM (n = 3). ∗∗∗p < 0.001; one-way ANOVA analysis.
(B and C) Expression levels of pluripotency genes in ΔPG (B) and ΔG (C) ESCs detected by qRT-PCR and western Blot. Three biological replicates per sample were assayed. Data are shown as the mean ± SEM (n = 3). ∗∗p < 0.01, ∗∗∗p < 0.001; one-way ANOVA analysis.
(D and E) Expression levels of differentiation genes in differentiated ΔPG (D) and ΔG (E) ESCs detected by qRT-PCR. ESCs are treated with retinoic acid (2 μM) for 4 days to differentiate. Three biological replicates per sample were assayed. Data are shown as the mean ± SEM (n = 3). ∗p < 0.05, ∗∗p < 0.01. Contrast in one-way ANOVA analysis was carried out (A–E).
(F and G) Relative size of EBs formed by ΔPG (F), ΔG (G), and WT ESCs. Three biological replicates per sample were assayed. Data are shown as the mean ± SEM, ∗∗∗p < 0.001; one-way ANOVA analysis. Scale bars, 100 μm.