Figure 3.
hPGCLCs in Extended Culture Self-Renew and Undergo Expansion
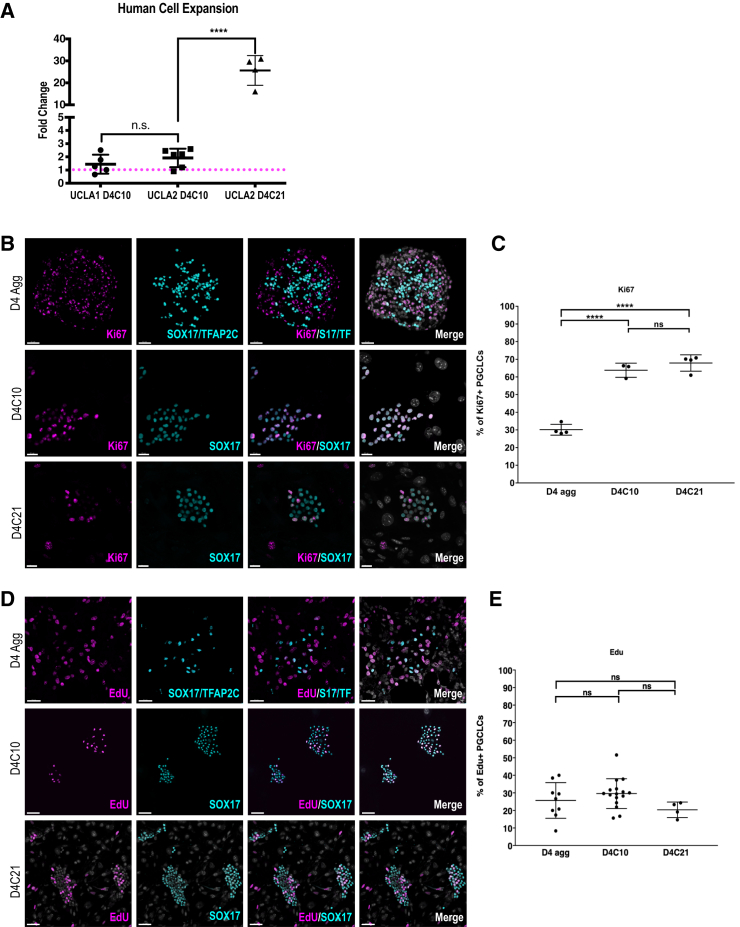
(A) Dot plot of the fold change in FACS isolated D4C10 and D4C21 hPGCLCs, compared with the starting D4 hPGCLCs. UCLA1 D4C10 (n = 5); UCLA2 D4C10 (n = 6) and D4C21 (n = 4). N = technical replicates. UCLA1 D4C10 and UCLA2 D4C10 represent 3 independent experiments, UCLA2 D4C21 represent 2 independent experiments. Magenta dotted line represents a fold change of 1. N.S., not significant; ∗∗∗∗p ≤ 0.0001. Error bars = mean SD.
(B) IF images of Ki67 (magenta) in UCLA2 D4 aggregate hPGCLCs, marked by SOX17/TFAP2C (cyan) (top panel), D4C10 hPGCLCs, marked by SOX17 (cyan) (middle panel), and D4C21 hPGCLCs, marked by SOX17 (cyan) (bottom panel). S17/TF = SOX17/TFAP2C. Scale bars, 50 μm (top), 20 μm (middle), and 20 μm (bottom).
(C) Quantification of Ki67-positive hPGCLCs in UCLA2 D4 aggregates (D4 agg), D4C10, and D4C21 hPGCLCs in FR10 media. ∗∗∗∗p ≤ 0.0001. Error bars = mean SD for D4 agg (N = 4), D4C10 (N = 3), and D4C21 (N = 3), N, the number of independent biological replicates.
(D) IF images of 5-ethynyl-2′-deoxyuridine (EdU) (magenta) in UCLA2 D4 aggregate hPGCLCs, marked by SOX17/TFAP2C (cyan) (top panel), D4C10 hPGCLCs, marked by SOX17 (cyan) (middle panel), and D4C21 hPGCLCs, marked by SOX17 (cyan) (bottom panel). S17/TF = SOX17/TFAP2C. Scale bars, 30 μm (top), 50 μm (middle), and 50 μm (bottom).
(E) Quantification of EdU-positive hPGCLCs in UCLA2 D4 aggregates (D4 agg), D4C10, and D4C21 hPGCLCs in FR10 media. N.S., not significant. Error bars = mean SD of 9 aggregates (D4 agg), 16 colonies (D4C10), and 4 colonies (D4C21) from 3 independent biological replicates.