Figure 5.
hPGCLCs in Extended Culture Undergo Partial Genome-Wide Demethylation
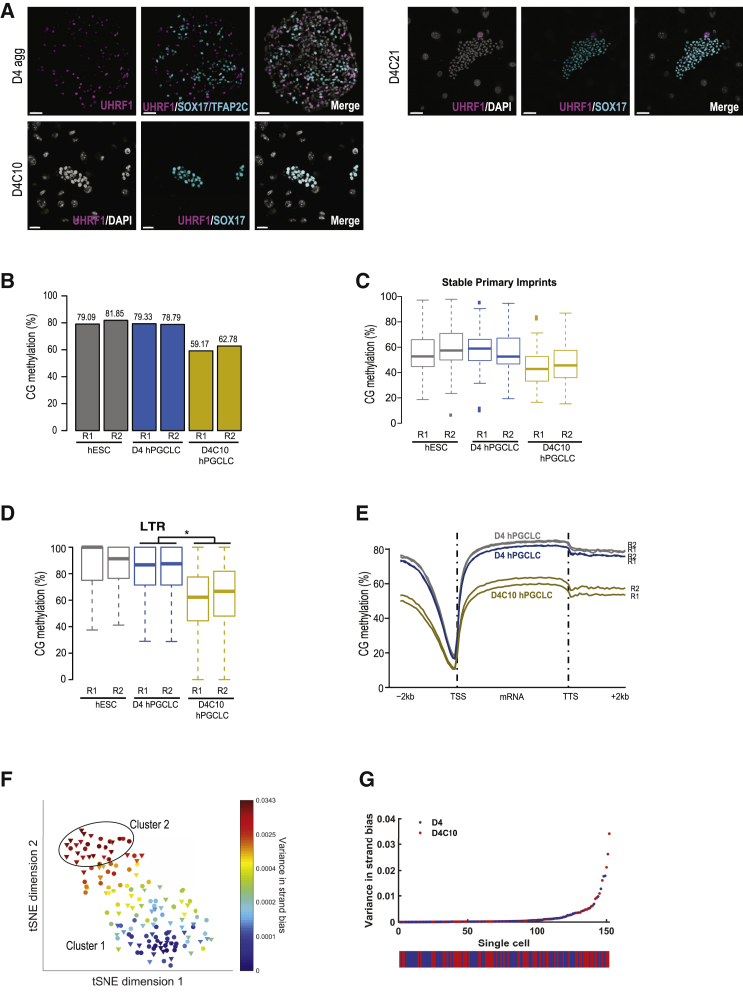
(A) IF images of UHRF1 expression in UCLA2 (U2) D4 aggregate hPGCLCs, marked by SOX17/TFAP2C (cyan) (top left panel), U2 D4C10 hPGCLCs, marked by SOX17 (cyan) (bottom left panel), and U2 D4C21 hPGCLCs, marked by SOX17 (cyan) (top right panel). Scale bars, 50 μm.
(B) Bar graph of average percent CG methylation in UCLA2: hESCs (gray), D4 hPGCLCs (blue), and D4C10 hPGCLCs (yellow).
(C) Boxplot of CG methylation percentages in U2 hESCs, D4 hPGCLCs, and D4C10 hPGCLCs over primary imprints (N = 31). U2 hESCs (gray), U2 D4 hPGCLCs (blue), and U2 D4C10 hPGCLCs (yellow).
(D) Boxplot of CG methylation percentages over long terminal repeats (LTRs). U2 hESCs (gray), U2 D4 hPGCLCs (blue), and U2 D4C10 hPGCLCs (yellow). ∗p < 2.2e16.
(E) Metaplot of percent CG methylation over protein coding genes and flanking 2-kb regions in U2 hESCs (gray), U2 D4 hPGCLCs (blue), and U2 D4C10 hPGCLCs (yellow). TSS, transcription start site; TTS, transcription termination site.
Replicates for (B–E) are independent replicates. R1, replicate 1; R2, replicate 2.
(F) t-SNE plot based of the strand bias of single-cell U2 D4 aggregate hPGCLCs (dots) and U2 D4C10 hPGCLCs cultured in FR10 (triangles). Cluster 1, low variance; cluster 2, high variance. Black circle indicate cells with the highest strand bias variance.
(G) Dot plot of each individual U2 D4 hPGCLC (blue) and U2 D4C10 hPGCLC (red), ordered by variance in strand bias.