FIGURE 1.
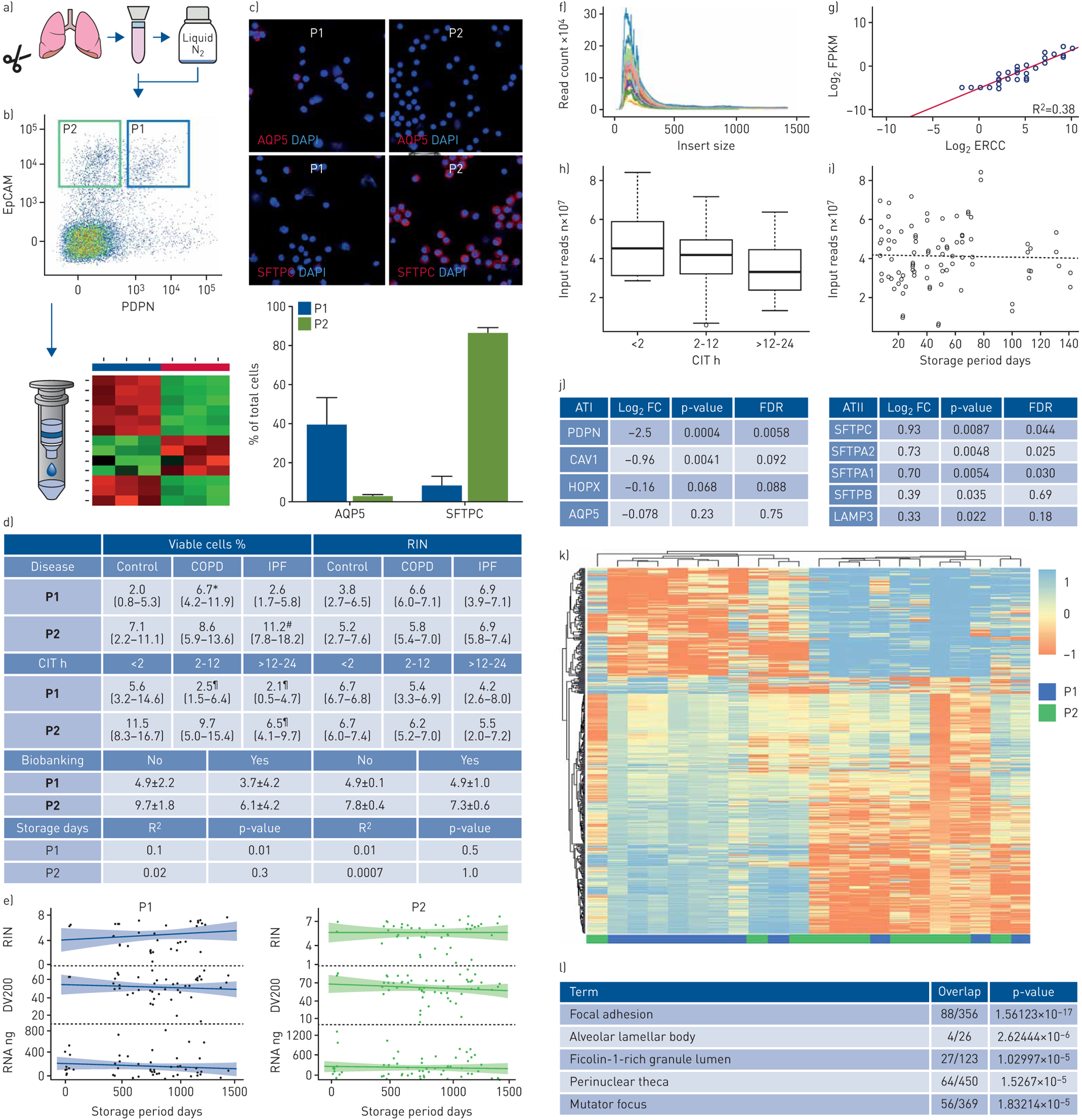
Isolation and RNA-seq of human alveolar epithelial cells (AECs). a) 52 human lung explants (20 control, 10 COPD and 22 idiopathic pulmonary fibrosis (IPF)) were enzymatically digested into cell suspensions that were biobanked in liquid nitrogen or flow-sorted immediately. b) Non-biobanked (n=6) and biobanked (n=52) cell suspensions from control and diseased lungs were flow-sorted using two gates, P1 and P2, to enrich for type I and II AECs (ATI and ATII), respectively. c) AQP5 and SFTPC expression in paired P1- and P2-gated biobanked control cells was quantified by immunofluorescence (n=3). d and e) Effects of cold ischaemic time (CIT), biobanking and storage duration (median 726 days, P25-P75 441–1021) on yield of viable P1- and P2-gated cells (as a percentage of total pre-sorted cells) and quality of RNA extracted from control and diseased lungs (n=52). RNA-seq was performed on paired P1- and P2-gated biobanked control samples (n=11). Quality control metrics demonstrated an adequate median insert size (f) and a high level of agreement between observed and expected concentrations of the target sequences (g), as well as a significant correlation of input size with CIT (h) but not storage duration (i) (p=0.03 and 0.79, respectively). j) Gene expression of canonical markers for ATI (left) and ATII (right) was significantly elevated in P1- and P2-gated cell populations, respectively, although not all comparisons reached statistical significance (e.g. for AQP5). Log2 FC reflects fold change of gene expression in P2- versus P1-gated samples. k) Hierarchical clustering of the top 650 genes revealed good separation of P1- and P2-gated samples from biobanked specimens. l) Pathway enrichment analysis with gene ontology terms was performed on differentially expressed P2-gated cells. *: p<0.05 versus control and IPF; #: p<0.05 versus control; ¶: p<0.05 versus <2 h. FC: fold change; FPKM: fragments per kilobase of transcript per million mapped reads; ERCC: external RNA control consortium spike-in; FDR: false discovery rate; RIN: RNA integrity number.
