Figure 1: Analysis strategy used to identify a new gene signature and train predictive models using Africa dataset and quantitatively test predictive performance in Brazil datasets.
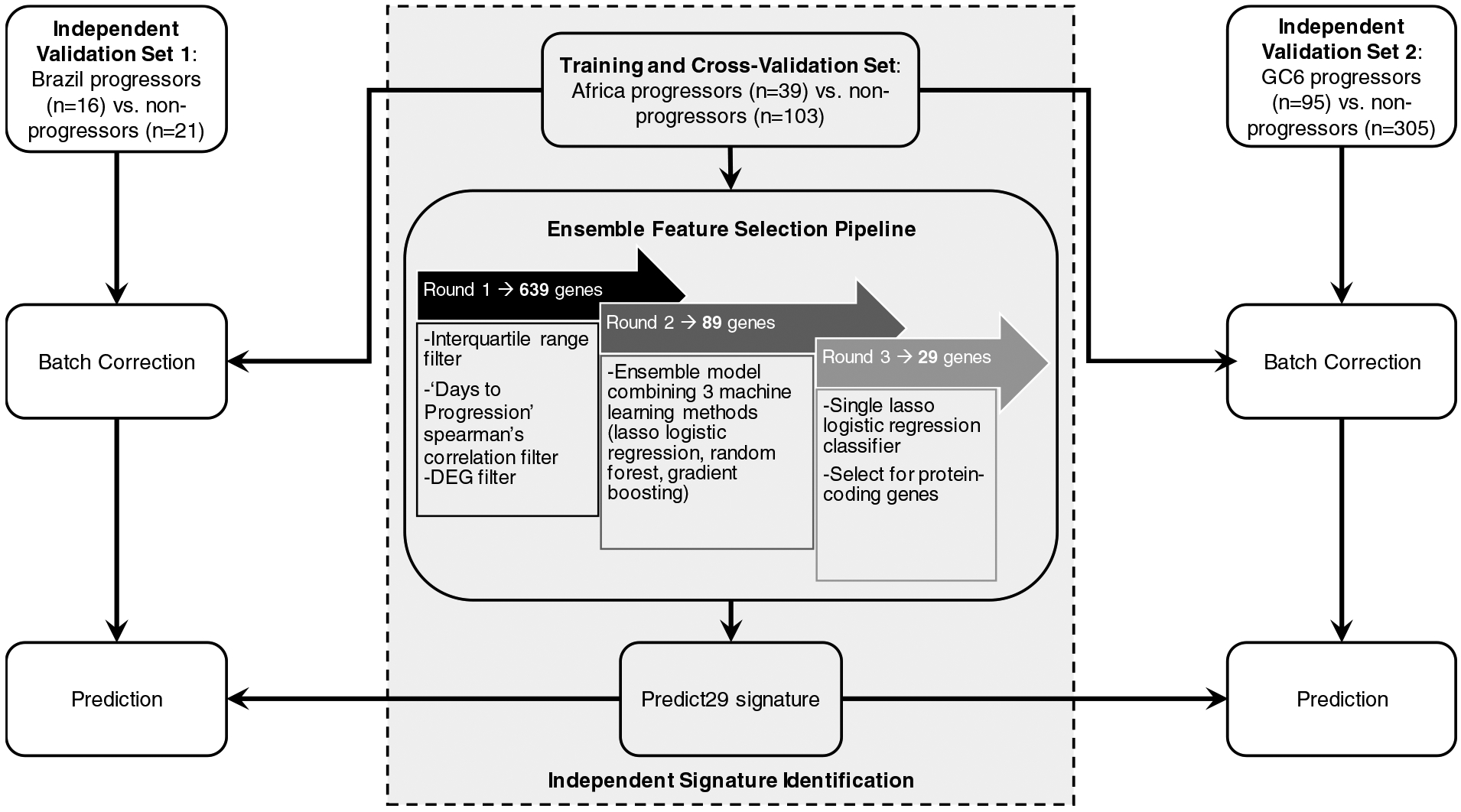
The Africa dataset (ACS-COR; GSE79362) derived from whole blood samples was used to identify a novel 29-gene signature via an ensemble feature selection pipeline: Round 1 led to identification of 639 genes of interest based on expression trends that correlated with progression. Round 2 led to selection of 89 genes based on evaluation using an ensemble model to determine which genes performed most robustly across different models. Round 3 led to final selection of 29-protein coding genes after removing redundant features. Predictive model training was performed using batch-corrected ACS-COR (GSE79362) Africa dataset (Training and Cross-Validation Set), and predictive testing was performed using batch-corrected Brazil progressors vs. non-progressor dataset derived from PBMC samples (Validation Set 1) and GC6-GSE94438 derived from whole blood samples (Validation Set 2).
