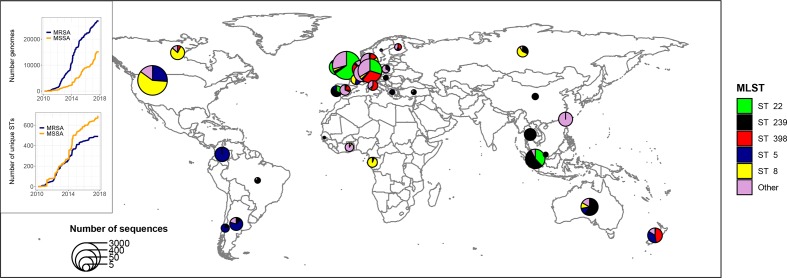
Fig. 1.
Snapshot of the genomic epidemiology of MRSA based on 42 948 publicly available S. aureus genomes (27 120 mec-positive) processed through the Staphopia platform (https://staphopia.emory.edu/). The inset shows the explosion of sequenced genomes and the constant increase in genetic diversity with 1099 different STs found in Staphopia. Despite this diversity, 70 % of MRSA sequences belonged to five STs (ST22, ST8, ST5, ST239 and ST398). It should be noted that publicly available S. aureus genomics do not accurately represent S. aureus epidemiology at this stage, due to sequencing and availability bias and lack of metadata in a large proportion of the dataset.