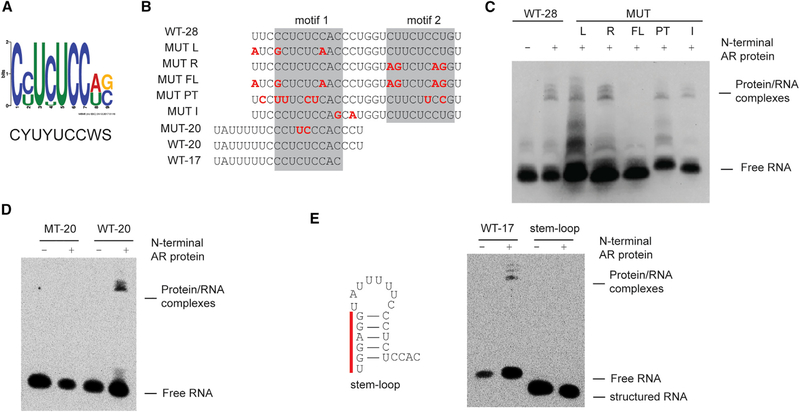
Figure 2. AR Binds to Single-Strand RNA in a Sequence-Specific Manner.
(A) Consensus motif identified among AR-binding RNA sequences.
(B) Sequences ofbiotinylated probes used in these assays. Nucleotidesthat are mutated compared towild-type SLNCR1 are highlighted in red, bolded font. The predicted AR-binding motifs are highlighted in the gray boxes.
(C and D) REMSA, as in Figure 1B, of the indicated probes derived from WT-28 (C) or WT-20 (D) incubated with 600 nM of recombinant AR NTD.
(E) REMSA, as in (C) and (D), with the indicated WT or stem-loop probe. Left: secondary structure model (ΔG = −5.4, as predicted by RNAStructure, rna.urmc.rochester.edu/RNAstructureWeb) ofstem-loop probewith the non-endogenous nucleotides denoted by the red bar. Significance was calculated using the Student’s t test: *p < 0.05; **p < 0.005; ns, not significant.
See also Figure S2.