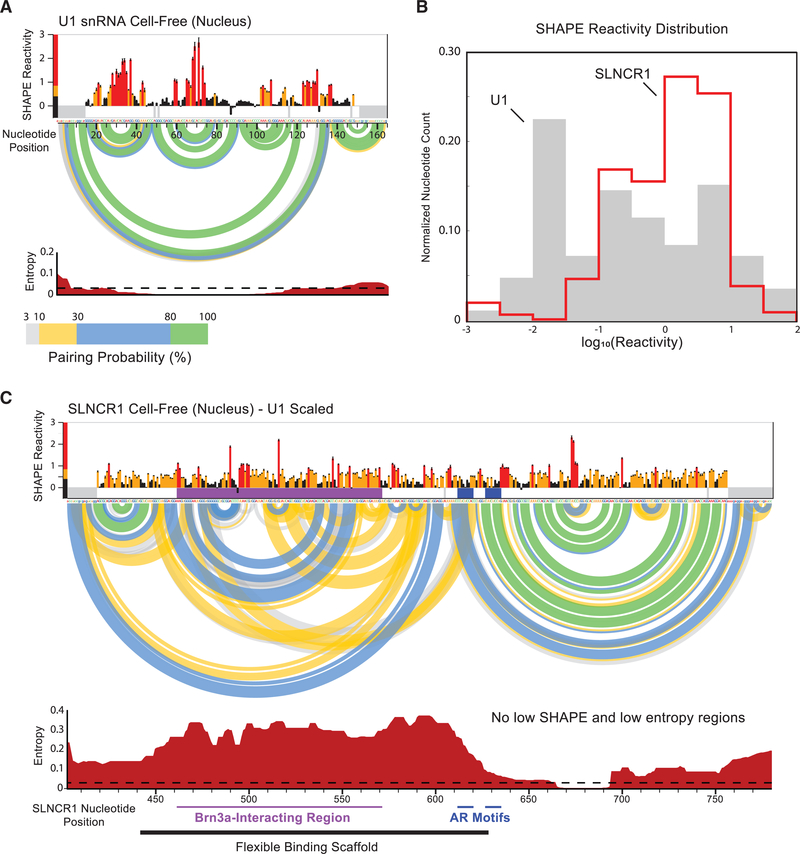
Figure 5. SHAPE-Directed RNA Structure Modeling Reveals SLNCR1403–780 is Largely Structured.
(A) Reactivity profile for the U1 small nuclear RNA(snRNA) from SHAPE-MaP of cell-free nuclear RNA. SHAPE reactivities are colored by relative value (see scale) and SE is indicated. The base-pairing probabilities of nucleotide pairs are represented by colored arcs linking the involved nucleotides. Entropy values (bottom) are local 50-nt windowed medians. A low entropy threshold consistent with limited conformational dynamics (equal to 0.03) is indicated with a dotted line. Nearly all nucleotides in U1 have windowed-median SHAPE and entropy values below thresholds suggestive of formation of well-defined RNA secondary structure (0.4 for SHAPE, 0.03 for entropy).
(B) Comparison of SHAPE reactivity distributions for SLNCR1 and U1 RNAs from nuclear extracts. 0nly nucleotides 403–780 of SLNCR1 were analyzed.
(C) Secondary structure model for nucleotides 403–780 of SLNCR1 lncRNA. SHAPE reactivities and Shannon entropies are plotted on the same scale as the U1 snRNA, shown in (A). Underlying SLNCR data are the same as shown in Figure 4A. See also Figure S6.