Abstract
Several genome engineering applications of CRISPR-Cas9, an RNA-guided DNA endonuclease, require precision control of Cas9 activity over dosage, timing, and targeted site in an organism. While some control of Cas9 activity over dose and time have been achieved using small molecules, and spatial control using light, no singular system with control over all the three attributes exists. Furthermore, the reported small-molecule systems lack wide-dynamic range, have background activity in absence of the small-molecule controller, and are not biologically inert, while the optogenetic systems require prolonged exposure of high-intensity light. We previously reported a small-molecule-controlled Cas9 system with some dosage and temporal control. By photocaging this Cas9 activator to render it biologically inert and photoactivatable, and employing next-generation protein engineering approaches, we have built a system with wide-dynamic range, low background, and fast photoactivation using a low-intensity light while rendering the small molecule activator biologically inert. We anticipate these precision controls will propel the development of practical applications of Cas9.
Keywords: CRISPR-Cas9, genome editing, transcription, photocaging, trimethoprim
Graphical Abstract
A singular system for precision control of Cas9 activity over dosage, timing and targeted site in an organism.
CRISPR-Cas9 is an RNA-guided DNA endonuclease[1] that has facilitated genome engineering applications in biotechnology, medicine, and agriculture. [2] These applications require precision control of Cas9 activity over multiple dimensions, including dose, temporal, and spatial.[3] Controlling the dose is particularly imsportant for Cas9, as the targeted DNA sequence is present at a much lower concentration than the enzyme, causing significant off-target effects,[3b, 4] chromosomal translocations,[4a, 5] and DNA-strand-break-triggered genotoxicity[6] at elevated Cas9 activity. Furthermore, as the off-target activity of Cas9 generally occurs at a slower rate than the on-target activity,[7] restricting the nuclease activity to a narrow temporal window is highly desired. Finally, several delivery systems accumulate Cas9 in specific off-target tissues,[8] so spatial restriction of Cas9 activity could avert off-target genotoxicity. Beyond genome editing, the fusion of catalytically-impaired Cas9 to effector domains has enabled targeted transcriptional activation or repression, base editing, epigenetic modification, and chromatin imaging. [3b, 9] Equipping these transformative tools with precision control over the aforementioned dimensions will also be highly desirable and propel many biological studies.
Controlling the dosage and timing of Cas9 activity has been accomplished using small-molecule-modulated systems,[3b] and spatial control with light-activated protein domains[10] or photoactivatable residues,[11] however, a singular system with built-in control over all three attributes does not exist. Unfortunately, a simple combination of the existing systems does not provide an adequate solution since the precision of such system is insufficient for several biotechnological and therapeutic applications.[2b, 2d, 3b, 12] For example, small-molecule-activated systems have low dynamic range of Cas9 activity and retain significant background activity even in the absence of the small-molecule modulator, preventing precise temporal control.[3b] As an additional drawback, the currently employed small-molecule activators (e.g., trimethoprim, hydroxytamoxifen) are not biologically inert and in some cases can adversely impact the organism’s microbiome. The precise control of Cas9 systems using light requires: (i) the rapid activation of Cas9 on timescale of minutes upon illumination with minimal background, (ii) low light intensities and short exposure times, (iii) reversible dosing with multi-wavelength control of Cas9 activity, and (iv) cost-effectiveness. Unfortunately, the reported photoactivatable Cas9 systems require long exposure times (often on the timescale of hours) and/or high light intensities (e.g., laser-based light source). Additionally, these systems are restricted to a narrow spectral range, are irreversible in certain cases, and can be expensive if photocaged RNA residues[11b] are employed. Therefore, while the reported systems display some of the aforementioned required characteristics, a singular system encompassing all of the desired attributes has not been reported.
We previously reported a small-molecule-controlled Cas9 system[4b] that exhibited dosage and temporal control by fusing Cas9 to the destabilized domains (DDs) of E. coli’s dihydrofolate reductase (DHFR).[13] These domains are unstable and rapidly target the fusion protein for proteasomal degradation, unless it is stabilized by the small molecule trimethoprim (TMP), thereby linking the activity of the fusion protein directly to the presence of TMP in a rapid, reversible, and dose-dependent manner (Fig. 1A). By controlling the concentration and time of administration or removal of TMP, both the dosage and temporal control of Cas9 activity was achieved. Though this system meets some of the requirements of the aforementioned singular system, it still lacks several others. First, the system exhibits significant background activity (~20%) in the absence of TMP, preventing precise temporal control of Cas9 activity. Second, the small molecule activator, TMP, is a potent anti-bacterial drug[14] that perturbs the organism’s microbiota,[15] limiting in vivo applications. Finally, this system lacks spatial control of Cas9 activity.
Figure 1.
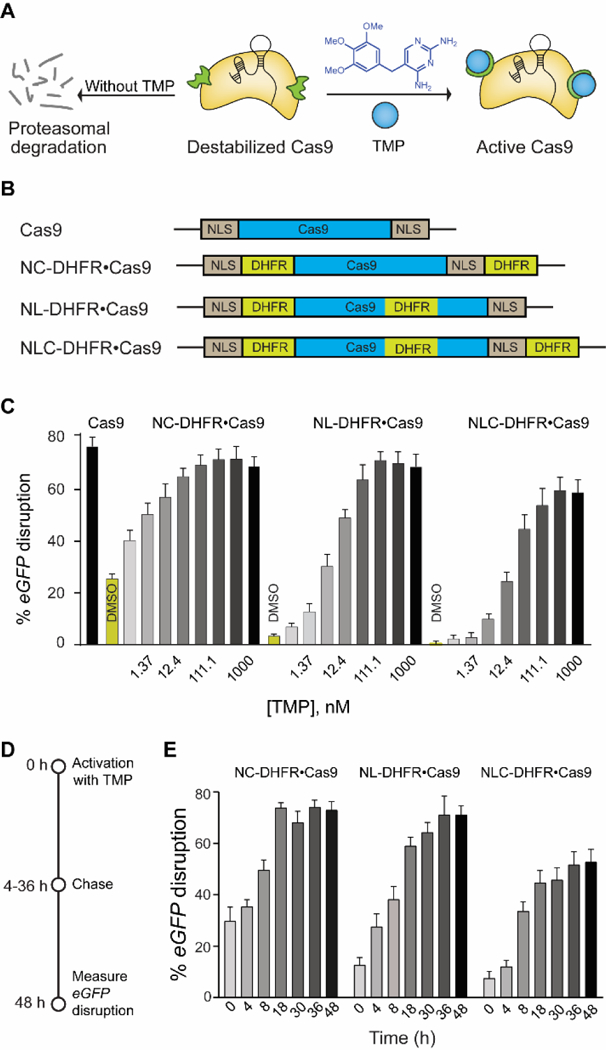
(A) Schematic representation of destabilized DHFR domains fused to Cas9, leading to proteosomal degradation. TMP stabilizes the DHFR•Cas9 fusion protein to generate active Cas9. (B) Schematic representation of different DHFR-fused Cas9 constructs. (C) Dose-dependent activation of different DHFR•Cas9 systems in the eGFP-disruption assay in U2OS cells (D) Schematic representation of reversible activation of Cas9. Cells were nucleofected with different DHFR•Cas9 constructs and treated with TMP (37 nM). The media was then removed and fresh media without TMP was added at different time intervals over 48 h, and the cells were imaged after 48 h. (E) Comparison of reversible and dose dependent activation of three different DHFR•Cas9 constructs by TMP (37 nM).
We hypothesized that next-generation protein engineering approaches could reduce the background activity in the absence of TMP. Furthermore, we hypothesized that photocaging TMP would both abrogate systemic anti-bacterial activity and also allow spatial control using light. Photocaged small molecules can be readily delivered to cells at much higher concentrations than those attainable for light-activated protein domains or photocaged residues, and small-molecule photocages that can be efficiently uncaged at various wavelengths are readily available. As such, we postulated that using a photocaged TMP would enable rapid Cas9 activation at low-light intensities and at multiple wavelengths. Herein, we report a Cas9 system that can be activated by a low-intensity light (non-laser based) of multiple wavelengths with exposure times of only minutes. This system has a low background in the absence of TMP, is dosable and reversible, and is non-toxic to both mammalian and bacterial cells. Finally, our system can be deployed for optochemically controlling transcription using catalytically inactive Cas9 fused to a transcription activator.
In our previously reported system (Fig. 1A), the Cas9 protein contained DHFR domains on both the N- and C-termini (DHFR•Cas9•DHFR), though still retained ~20% background activity in the absence of TMP.[4b] In parallel, Savage and co-workers[16] identified loops on Cas9 that tolerated the insertion of protein domains without a loss of activity. We therefore hypothesized that grafting an additional DHFR domain on the Cas9 loop itself (termed “L-DHFR”) should further reduce the background activity in the absence of TMP (Fig. 1B). We tested this 3DHFR construct (Fig. 1B) in an eGFP-disruption assay[4b] in which Cas9-mediated eGFP-knockout causes loss of fluorescence. We observed background reduction by 5–7 fold and dose-dependent activation of Cas9, though the dynamic range was also reduced, with Cas9 exhibiting only 62% eGFP-disruption at a full-dose of TMP (Fig. 1C). Given the pronounced effect of the L-DHFR on background reduction, we hypothesized that systematically removing DHFR domains at the other sites (N-term, C-term, or both) of 3DHFR should yield a construct with a large enough dynamic range and minimal background activity. Indeed, the construct containing both an N-DHFR and L-DHFR domain had the minimal background and retained a dynamic range similar to that of wildtype Cas9 (Fig. 1C). Next, we confirmed the reversible nature of TMP-mediated activation in a “washout” experiment (Fig. 1D, E). Briefly, the cells were nucleofected with the various destabilized domain constructs and treated with TMP. The media was then exchanged for fresh media without TMP at different time intervals over 48 h (Fig. 1D). The cells exposed to TMP for longer durations showed a higher eGFP-disruption and Cas9 activity, and such reversibility was observed for a wide range of TMP concentrations from 4 nM to 1 μM (Fig. S1).
After developing this improved inducible Cas9 system with minimal background activity, we proceeded to introduce optical control and abrogate the anti-bacterial activity of TMP that can adversely impact the microbiome.[15] We hypothesized that appending photocleavable moieties to TMP would render it inactive and that exposure to light would remove the photocage to release the free activator. Before embarking on any photocaging experiments, we confirmed that TMP was photostable at various wavelengths and light intensities. Indeed, no functional modification or detectable degradation of TMP was observed via liquid chromatography-mass spectrometry across the range of wavelengths and intensities that were relevant for our photocages (Fig. S2).
To design photocaged TMP, we examined the co-crystal structure of TMP bound to DHFR (Fig. 2A) showing that both aromatic amino groups of TMP are buried inside the binding pocket of the DHFR. A bulky photocaging group installed on these amino groups should therefore adversely impact the ability of TMP to bind to and destabilize DHFR. We explored two types of photocaging moieties (Fig. 2B) reported for their use in the precise photoregulation of various biological processes.[17] For example, photocaging group 2-(2-nitrophenyl)-propoxycarbonyl (NPPOC) has been used for spatiotemporal control of cells but long exposures to wavelengths employed for NPPOC uncaging can induce genomic lesions,[18] and the tissue penetration of such wavelengths is also limited. As such, we decided to employ the photocaging group thiocoumarine, which can be removed by irradiation with visible light that is safer.[19] We synthesized the photocaged TMP compounds 1 and 2 (Fig. 2B), which are uncaged at wavelengths of 385 nm and 470 nm, respectively. We confirmed that photocaged TMP did not prevent bacterial growth, supporting our hypothesis that the photocaged molecule would not adversely impact the microbiome as done by TMP (Fig. S3). Exposure of the photocaged TMP to light removed the photocaging moiety and released active TMP (Fig. S4). We have used the LED device previously reported[20] by some of us for our experiments.
Figure 2.
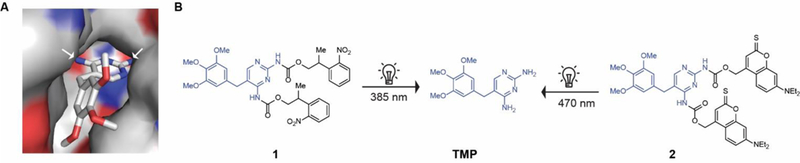
(A) Crystal structure of the TMP-DHFR complex showing that the NH2 groups of TMP are deeply buried inside the binding pocket. (B) Chemical structure of the photocaged TMP derivatives 1 and 2 and unmasking of photocaging group under UV and visible light, respectively.
Next, we used compound 1 or 2 to control DHFR-based Cas9 systems using light in the same eGFP-disruption assay that was used to quantify Cas9 activity. The cells were nucleofected with different destabilized domain constructs along with the eGFP-targeting gRNA plasmid and then incubated with compound 1 (Fig. 3A, S7A) or 2 (Fig. 3E, S8A). There was no noticeable loss of eGFP signal in the absence of light exposure, even after 48 h, though exposure to light of either 385 nm or 470 nm for only a few minutes caused a dose-dependent eGFP-knockdown that would be expected for a successful photoactivation of a caged molecule (Fig. 3B, 3F, S7B, S8B). Next, we varied the light exposure time using a fixed concentration of the photocaged TMP, and we found that the extent of eGFP-knockdown was proportional to the light exposure time for both compound 1 and 2 (Fig. 3C, 3G, S7C, S8C). Both compounds showed a two-dimensional activation control that could be achieved by either varying the concentration of the photocaged TMP or the duration of light exposure (Fig. S7, 8). We confirmed comparable activation of Cas9 when cells were first treated with compound 1 or 2 that was then removed through media swap before light exposure (Fig. S9, 10). In addition to short exposure times of 12 minutes, our system employs low intensity LED lights with the low power (8 mW/cm2 for 385 nm and 14.7 mW/cm2 for 470 nm LEDs). This low light intensity and miniature light source will allow facile optical control of Cas9 in vivo using various devices, including optical fibers[21] or implantable wireless photonic devices.[22] We were also able to conduct the experiments in a 96-well plate by placing a photomask patterned to spell “CAS” and observed eGFP-disruption in only the light exposed wells of the microplate, resulting in a spelled-out “CAS” pattern (Fig. S11).
Figure 3:
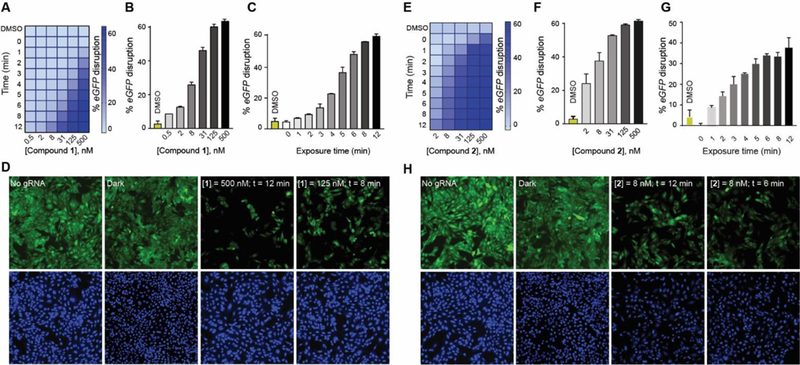
(A) Two-dimensional photo-chemical activation of NL-DHFR•Cas9 activity in the eGFP-disruption assay in U2OS cells. Cells nucleofected with NL-DHFR•Cas9 and eGFP-targeting gRNA plasmids were incubated with at various concentrations of compound 1 and exposed to increasing amount of UVA light followed by 48 h of incubation. (B) Dose-dependent photoactivation of the NL-DHFR•Cas9 system with compound 1 in the eGFP-disruption assay in U2OS cells upon exposure to 12 min of UVA light (C) UVA light exposure time-dependent photoactivation of the NL-DHFR•Cas9 system in the eGFP-disruption assay in U2OS cells in the presence of 125 nM of compound 1. (D) Representative images of the eGFP-disruption assay by NL-DHFR•Cas9 under various conditions: no gRNA, dark, 500 nM of compound 1 and 12 min of UVA exposure, or 8 min UVA exposure and 125 nM of compound 1. (E) Two-dimensional photochemical activation of NL-DHFR•Cas9 by compound 2 and visible light. (F) Dose-dependent photoactivation of the NL-DHFR•Cas9 system with compound 2 in the eGFP-disruption assay upon exposure to 12 min of visible light. (G) Visible-light-dependent photoactivation of the NL-DHFR•Cas9 system in the eGFP-disruption assay in U2OS cells in the presence of 8 nM of compound 2. (H) Representative images of the eGFP-disruption assay by NL-DHFR•Cas9 under various conditions: no gRNA, dark, 8 nM of compound 2 and 12 min of visible light exposure, or 6 min of visible light exposure and 8 nM of compound 2.
We next investigated the utility of our system to control catalytically-dead Cas9 (dCas9) for precision optochemical control of transcriptional activation. We used our previously reported[4b] transcriptional activation system (DHFR•PP7•VP64) consisting of transcriptional activation domain VP64 and PP7, which binds to a specific sequence displayed on the gRNA. Upon treatment with varying doses of TMP, we observed robust and dose-dependent upregulation of IL1RN as well as minimal basal activity in the absence of TMP (Fig. S12). We then tested the optical control of IL1RN transcription using the DHFR•PP7•VP64 system with various doses of compound 1. Upon light illumination for 12 minutes, we observed upregulation of IL1RN transcription proportional to the dose of the compound (Fig. S13A). Further, to investigate the effect of light exposure time on transcriptional upregulation, we treated cells transfected with dCas9, DHFR•PP7•VP64, and gRNA with 500 nM of 1 and exposed them to 385 nm of light for varying periods (Fig. S13B). We observed that the upregulation of IL1RN was proportional to the duration of light exposure. Notably, we observed minimal basal activation of IL1RN transcription in the absence of either the compound 1 or light exposure (Fig. S13C). Similarly, we were able to control the transcription of IL1RN by varying the concentration of compound 2 or varying the exposure to visible light (Fig. S14). These studies suggest that our optochemical system can be used for precision control of both catalytically-active Cas9 and technologies based on catalytically-impaired Cas9. Finally, we note that the transcriptional activation system based on those reported by Dahlman et al.[23] failed to provide an optochemical system with low background and high dynamic range (Fig. S15-17).
In summary, we developed a singular system with dose, temporal, and spatial control over Cas9 activity. This system possesses a low background activity in the absence of TMP and has an activity comparable to wild type Cas9 at a full dose of TMP. Furthermore, this system is dosable, reversible, non-toxic to both mammalian and bacterial cells, and afford precision optical control of Cas9 activity by allowing photoactivation in minutes using low-intensity light. Our system can be deployed for optochemical control of transcription using catalytically inactive Cas9 fused to a transcriptional activator. Finally, our approach is extendable to other reported small-molecule regulated Cas9 systems. These studies will propel the development of therapeutic and biotechnological applications of CRISPR-Cas9.
Supplementary Material
Acknowledgements
This work was supported by the Burroughs Wellcome Fund (Career Award at the Scientific Interface), DARPA (N66001-17-2-4055), NIH (R21AI126239), and Army Research Office Award W911NF1610586.
This work is dedicated to Professor Ronald T. Raines on the occasion of his 60th birthday.
Footnotes
Conflict of interest
Broad Institute has filed a patent application including work from this paper
References
- [1].a) Jiang F, Doudna JA, Annu. Rev. Biophys. 2017, 46, 505–529; [DOI] [PubMed] [Google Scholar]; b) Chen JS, Doudna JA, Nat. Rev. Chem. 2017, 1, 1–15. [Google Scholar]
- [2].a) Hsu PD, Lander ES, Zhang F, Cell 2014, 157, 1262–1278; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Cox DBT, Platt RJ, Zhang F, Nat. Med. 2015, 21, 121–131; [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Bryson TE, Anglin CM, Bridges PH, Cottle RN, Yale J Biol. Med. 2017, 90, 553–566; [PMC free article] [PubMed] [Google Scholar]; d) Fellmann C, Cowen BC, Lin PC, Doudna JA, Corn JE, Nat. Rev. Drug. Discov. 2017, 16, 89–100; [DOI] [PMC free article] [PubMed] [Google Scholar]; e) Gao CX, Nat. Rev. Mol. Cell. Bio. 2018, 19, 275–276. [DOI] [PubMed] [Google Scholar]
- [3].a) Nunez JK, Harrington LB, Doudna JA, Acs. Chem. Biol. 2016, 11, 681–688; [DOI] [PubMed] [Google Scholar]; b) Gangopadhyay SA, Cox KJ, Manna D, Lim D, Maji B, Zhou Q, Choudhary A, Biochemistry 2019, 58, 234–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].a) Frock RL, Hu J, Meyers RM, Ho YJ, Kii E, Alt FW, Nat. Biotechnol. 2015, 33, 179–186; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Maji B, Moore CL, Zetsche B, Volz SE, Zhang F, Shoulders MD, Choudhary A, Nat. Chem. Biol. 2017, 13, 9–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Vanoli F, Tomishima M, Feng WR, Lamribet K, Babin L, Brunet E, Jasin M, Proc. Natl. Acad. Sci. U.S.A. 2017, 114, 3696–3701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].a) Ihry RJ, Worringer KA, Salick MR, Frias E, Ho D, Theriault K, Kommineni S, Chen J, Sondey M, Ye CY, Randhawa R, Kulkarni T, Yang Z, McAllister G, Russ C, Reece-Hoyes J, Forrester W, Hoffman GR, Dolmetsch R, Kaykas A, Nat. Med. 2018, 24, 939–946; [DOI] [PubMed] [Google Scholar]; b) Haapaniemi E, Botla S, Persson J, Schmierer B, Taipale J, Nat. Med. 2018, 24, 927–930. [DOI] [PubMed] [Google Scholar]
- [7].Shin J, Jiang FG, Liu JJ, Bray NL, Rauch BJ, Baik SH, Nogales E, Bondy-Denomy J, Corn JE, Doudna JA, Sci. Adv. 2017, 3, 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Li C, Psatha N, Gil S, Wang HJ, Papayannopoulou T, Lieber A, Mol. Ther. Methods Clin. Dev. 2018, 9, 390–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].a) Rees HA, Liu DR, Nat. Rev. Genet. 2018, 19, 770–788; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Lo A, Qi L, F1000Research 2017, 6; [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Wang HF, La Russa M, Qi LS, Annu. Rev. Biochem. 2016, 85, 227–264; [DOI] [PubMed] [Google Scholar]; d) Komor AC, Badran AH, Liu DR, Cell 2017, 168, 20–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].a) Nihongaki Y, Kawano F, Nakajima T, Sato M, Nat. Biotechnol. 2015, 33, 755–760; [DOI] [PubMed] [Google Scholar]; b) Nihongaki Y, Yamamoto S, Kawano F, Suzuki H, Sato M, Chem. Biol. 2015, 22, 169–174; [DOI] [PubMed] [Google Scholar]; c) Nihongaki Y, Furuhata Y, Otabe T, Hasegawa S, Yoshimoto K, Sato M, Nat. Methods 2017, 14, 963–966; [DOI] [PubMed] [Google Scholar]; d) Zhou XX, Zou XZ, Chung HYK, Gao YC, Liu YX, Qi LS, Lin MZ, Acs. Chem. Biol. 2018, 13, 443–448; [DOI] [PMC free article] [PubMed] [Google Scholar]; e) Polstein LR, Gersbach CA, Nat. Chem. Biol. 2015, 11, 198–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].a) Hemphill J, Borchardt EK, Brown K, Asokan A, Deiters A, J. Am. Chem. Soc. 2015, 137, 5642–5645; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Jain PK, Ramanan V, Schepers AG, Dalvie NS, Panda A, Fleming HE, Bhatia SN, Angew. Chem. Int. Ed. 2016, 55, 12440–12444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Dunbar CE, High KA, Joung JK, Kohn DB, Ozawa K, Sadelain M, Science 2018, 359, 1–10. [DOI] [PubMed] [Google Scholar]
- [13].a) Miyazaki Y, Imoto H, Chen LC, Wandless TJ, J. Am. Chem. Soc. 2012, 134, 3942–3945; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Banaszynski LA, Chen LC, Maynard-Smith LA, Ooi AGL, Wandless TJ, Cell 2006, 126, 995–1004; [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Iwamoto M, Bjorklund T, Lundberg C, Kirik D, Wandless TJ, Chem. Biol. 2010, 17, 981–988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Bushby SR, Hitchings GH, Br. J. Pharmaco.l Chemother. 1968, 33, 72–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Modi SR, Collins JJ, Relman DA, J. Clin. Invest. 2014, 124, 4212–4218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Oakes BL, Nadler DC, Flamholz A, Fellmann C, Staahl BT, Doudna JA, Savage DF, Nat. Biotechnol. 2016, 34, 646–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].a) Hansen MJ, Velema WA, Lerch MM, Szymanski W, Feringa BL, Chem. Soc. Rev. 2015, 44, 3358–3377; [DOI] [PubMed] [Google Scholar]; b) Klan P, Solomek T, Bochet CG, Blanc A, Givens R, Rubina M, Popik V, Kostikov A, Wirz J, Chem. Rev. 2013, 113, 119–191; [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Ankenbruck N, Courtney T, Naro Y, Deiters A, Angew. Chem. Int. Ed. 2018, 57, 2768–2798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Rastogi RP, Richa, Kumar A, Tyagi MB, Sinha RP, J. Nucleic Acids 2010, 2010, 1–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Fonseca ASC, Soares AMS, Goncalves MST, Costa SPG, Tetrahedron 2012, 68, 7892–7900. [Google Scholar]
- [20].Reis SA, Ghosh B, Hendricks JA, Szantai-Kis DM, Tork L, Ross KN, Lamb J, Read-Button W, Zheng BX, Wang HT, Salthouse C, Haggarty SJ, Mazitschek R, Nat. Chem. Biol. 2016, 12, 317–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].McCall JG, Kim TI, Shin G, Huang X, Jung YH, Al-Hasani R, Omenetto FG, Bruchas MR, Rogers JA, Nat. Protoc. 2013, 8, 2413–2428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Bansal A, Yang FY, Xi T, Zhang Y, Ho JS, Proc. Natl. Acad. Sci. U.S.A. 2018, 115, 1469–1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Dahlman JE, Abudayyeh OO, Joung J, Gootenberg JS, Zhang F, Konermann S, Nat. Biotechnol. 2015, 33, 1159–1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


