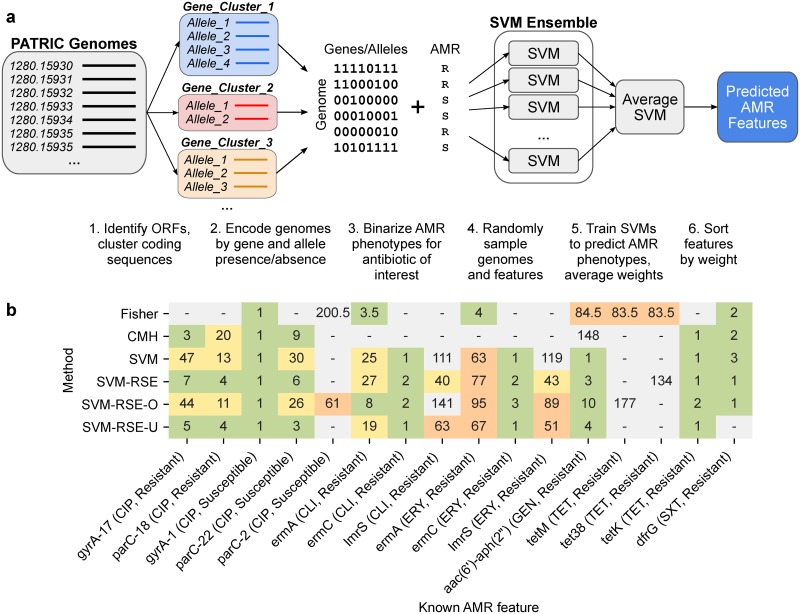
Fig 2. Comparison of SVM ensemble approaches and statistical tests for detecting AMR-conferring genes and alleles in S. aureus.
(a) Workflow for SVM ensemble approaches. Beginning with genomes from PATRIC, open reading frames (ORFs) are identified and clustered by coding sequence to identify putative genes and alleles. Each genome is encoded based on the presence or absence of each gene and allele to capture genomic variation in the pan-genome as a sparse binary matrix. Genomes and/or features of this matrix are randomly sampled 500 times and used to train SVMs to predict binary AMR phenotype for a single antibiotic from genotype. Weights for each feature are averaged across all models in the ensemble and used to rank features by association to AMR. (b) Associations between known AMR-conferring genomic features and AMR phenotype, as ranked by Fisher’s Exact test, Cochran-Mantel-Haenszel test, and four different SVM ensemble types (SVM: ensemble by bootstrapping genomes, SVM-RSE: bootstrapping genomes and features; “random subspace ensemble”, SVM-RSE-O: SVM-RSE with oversampling to balance subtypes, SVM-RSE-U: SVM-RSE with undersampling to balance subtypes). Features were ranked either by p-value for statistical tests or by average feature weight for SVM ensembles. Fractional ranking was used for ties. Only features detected by at least one method are shown, colored by rank (green: in top 10, yellow: 11–50, orange: 51–100, gray: >100). Features shown are either genes or individual alleles (denoted as <gene>-#).