Abstract
The incidence of oropharyngeal squamous cell carcinoma (OPSCC) has been rapidly increasing. Disease stage and smoking history are often used in current clinical trials to select patients for deintensification therapy, but these features lack sufficient accuracy for predicting disease relapse. Our purpose was to develop an imaging signature to assess early response and predict outcomes of OPSCC. Methods: We retrospectively analyzed 162 OPSCC patients treated with concurrent chemoradiotherapy, equally divided into separate training and validation cohorts with similar clinical characteristics. A robust consensus clustering approach was used to spatially partition the primary tumor and involved lymph nodes into subregions (i.e., habitats) based on 18F-FDG PET and contrast CT imaging. We proposed quantitative image features to characterize the temporal volumetric change of the habitats and peritumoral/nodal tissue between baseline and midtreatment. The reproducibility of these features was evaluated. We developed an imaging signature to predict progression-free survival (PFS) by fitting an L1-regularized Cox regression model. Results: We identified 3 phenotypically distinct intratumoral habitats: metabolically active and heterogeneous, enhancing and heterogeneous, and metabolically inactive and homogeneous. The final Cox model consisted of 4 habitat evolution-based features. In both cohorts, this imaging signature significantly outperformed traditional imaging metrics, including midtreatment metabolic tumor volume for predicting PFS, with a C-index of 0.72 versus 0.67 (training) and 0.66 versus 0.56 (validation). The imaging signature stratified patients into high-risk versus low-risk groups with 2-y PFS rates of 59.1% versus 89.4% (hazard ratio, 4.4; 95% confidence interval, 1.4–13.4 [training]) and 61.4% versus 87.8% (hazard ratio, 4.6; 95% confidence interval, 1.7–12.1 [validation]). The imaging signature remained an independent predictor of PFS in multivariable analysis adjusting for stage, human papillomavirus status, and smoking history. Conclusion: The proposed imaging signature allows more accurate prediction of disease progression and, if prospectively validated, may refine OPSCC patient selection for risk-adaptive therapy.
Keywords: prognostic biomarker, intratumoral subregion, peritumoral interaction, habitat evolution, early response
There has been a rapid increase in the prevalence of oropharyngeal squamous cell carcinoma (OPSCC), largely attributable to the epidemic of oral human papillomavirus (HPV) infection (1). The current standard treatment for locally advanced OPSCC is concurrent chemoradiotherapy, which is associated with significant morbidity and decreased quality of life. HPV-positive tumors typically respond well to radiation and chemotherapy, and these patients tend to have a better prognosis than those with HPV-negative tumors (2). Current treatment for HPV-positive OPSCC therefore likely represents overtreatment for many patients.
Currently, there are ongoing and proposed trials that focus on treatment deescalation in patients with clinically favorable HPV-positive OPSCC based on tumor/nodal stage and smoking history (3,4). These clinical features are imperfect, however, and some patients still develop progressive disease despite having a small tumor. In a recently reported phase III randomized trial (Radiation Therapy Oncology Group trial 1016), 987 patients were enrolled on the basis of clinical stage. This trial unfortunately failed to meet its primary objective of demonstrating the noninferiority of a less toxic drug (cetuximab) versus high-dose cisplatin, which is the current standard systemic treatment for this disease (5). This result highlights the unmet need for better prognostic biomarkers to improve risk stratification.
Imaging with CT and 18F-FDG PET is often used in staging, radiation treatment planning, and response evaluation for patients with OPSCC (6,7). Numerous studies have investigated imaging characteristics associated with treatment outcomes, with limited success (8–18). Most previous studies, however, analyzed baseline imaging before the initiation of therapy. This approach does not take into account the response to treatment, which can vary significantly across tumors. Another limitation is that most studies focus on the primary tumor and ignore regional lymph nodes, which are a frequent site for metastasis in OPSCC.
At our institution, we have been routinely acquiring contrast-enhanced CT and 18F-FDG PET scans both before and during the course of radiation treatment for OPSCC. The midtreatment scan is acquired for radiation replanning purposes, to address potential tumor response. An important advantage of midtreatment imaging is that it allows the assessment of early treatment response, which may be better correlated with disease outcomes (19,20).
In this work, we investigated the evolution of novel spatiotemporal imaging characteristics in intratumor subregions (i.e., habitats) to assess early response and predict treatment outcomes in OPSCC patients. We hypothesized that integrating quantitative image features of the primary tumor and lymph nodes extracted from baseline and midtreatment imaging will provide a more complete evaluation of the complex response patterns and may allow better prediction of clinical outcomes.
MATERIALS AND METHODS
Study Design
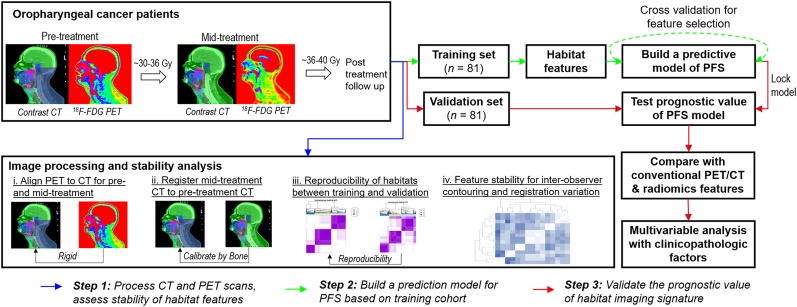
The overall study design is shown in Figure 1. In brief, we retrospectively collected and analyzed the baseline and midtreatment 18F-FDG PET and CT scans for patients with OPSCC who were treated with concurrent chemoradiotherapy. We proposed novel spatiotemporal habitat evolution-based image features to characterize the treatment response in the primary tumor and involved nodes, and we assessed the stability and reproducibility of extracted features. Next, we developed an imaging signature to predict PFS in a training cohort and evaluated it in an independent validation cohort. Finally, we assessed its independent prognostic value in relation to established imaging markers and clinicopathologic factors for predicting PFS.
FIGURE 1.
Proposed study design.
Patient Cohort
In this institutional review board–approved, Health Insurance Portability and Accountability Act–compliant study, we retrospectively collected data for patients with OPSCC who were consecutively treated at Stanford University Medical Center from February 2009 to February 2017. In total, 162 oropharyngeal patients were eligible for this study (Supplemental Fig. 1; supplemental materials are available at http://jnm.snmjournals.org).
Patients were included if they underwent definitive concurrent chemoradiotherapy for OPSCC; if pre- and midtreatment contrast-enhanced CT scans, 18F-FDG PET scans, and radiation treatment plans were available; and if they had been closely followed with posttreatment physical examination and PET/CT or MRI at a 3- to 4-mo interval after treatment completion. Patients were excluded if they did not have biopsy-confirmed squamous cell carcinoma, had previously received definitive surgery, had metastatic disease at presentation, or had no primary tumor (T0).
The overall study population was equally divided into separate training and validation cohorts. To mitigate random effects and ensure balanced splitting, we stratified the patients by matching T stage, N stage, HPV status, and smoking history. The demographic and clinical characteristics of the study population are presented in Table 1, which show similar distributions between the training and validation cohorts.
TABLE 1.
Demographic and Clinical Characteristics of Study Cohort
| Total (n = 162) | Training (n = 81) | Validation (n = 81) | P | |
| Median age (y) | 62.4 (SD, 9.8) | 64.3 (SD, 11.2) | 62.0 (SD, 8.2) | 0.138 |
| Sex (n) | 1.0 | |||
| Male | 149 (92.0%) | 74 (91.4%) | 75 (92.6%) | |
| Female | 13 (8.0%) | 7 (8.6%) | 6 (7.4%) | |
| T category (AJCC 7th ed.) (n) | 0.987 | |||
| T1 | 20 (12.3%) | 10 (12.3%) | 10 (12.3%) | |
| T2 | 58 (35.8%) | 30 (37.0%) | 28 (34.6%) | |
| T3 | 46 (28.4%) | 23 (28.4%) | 23 (28.4%) | |
| T4 | 38 (23.5%) | 18 (22.2%) | 20 (24.7%) | |
| N category (AJCC 7th ed.) (n) | 0.483 | |||
| N0 | 8 (4.9%) | 6 (7.4%) | 2 (2.5%) | |
| N1 | 15 (9.3%) | 7 (8.6%) | 8 (9.9%) | |
| N2 | 125 (77.2%) | 60 (74.1%) | 65 (80.2%) | |
| N3 | 14 (8.6%) | 8 (9.9%) | 6 (7.4%) | |
| Stage (AJCC 7th ed.) (n) | 1.0 | |||
| I | 1 (0.6%) | 1 (1.2%) | 0 (0%) | |
| II | 1 (0.6%) | 0 (0%) | 1 (1.2%) | |
| III | 13 (8.0%) | 7 (8.6%) | 6 (7.4%) | |
| IV | 147 (90.7%) | 73 (90.1%) | 74 (91.4%) | |
| p16 status (HPV) (n) | 0.812 | |||
| + | 142 (87.7%) | 70 (86.4%) | 72 (88.%9) | |
| − | 20 (12.3%) | 11 (13.6%) | 9 (11.1%) | |
| Smoking history (n) | 1.0 | |||
| >10 pack-years | 60 (37.0%) | 30 (37.0%) | 30 (37.0%) | |
| ≤10 pack-years | 102 (63.0%) | 51 (63.0%) | 51 (63.0%) | |
| PFS (n) | 1.0 | |||
| No | 37 (22.8%) | 18 (22.2%) | 19 (23.5%) | |
| Yes | 125 (77.2%) | 63 (77.8%) | 62 (76.5%) | |
| Median follow-up (y) | 2.1 (SD, 1.3) | 2.1 (SD, 1.4) | 2.1 (SD, 1.3) | 1.0 |
| 2-y PFS (n) | 1.0 | |||
| No | 32 (19.8%) | 16 (19.8%) | 16 (19.8%) | |
| Yes | 130 (80.2%) | 65 (80.2%) | 65 (80.2%) |
Imaging Protocol
All patients underwent an initial 18F-FDG PET scan and separate contrast-enhanced CT with a standard imaging protocol for radiation therapy planning. To account for potential tumor response, patients underwent a second 18F-FDG PET scan and separate contrast-enhanced CT for radiation therapy replanning after receiving approximately 30–36 Gy.
We analyzed the PET component of the 18F-FDG PET/CT scan, as well as a separate treatment-planning CT scan that had been obtained with contrast medium (not the CT component of the PET/CT scan). Two types of scanners were used: a GE Healthcare Discovery ST and a Siemens Biograph. Regarding the imaging protocol, contrast-enhanced CT scans were acquired at a tube potential of 120 kV and a tube current of 250 mA. CT images had a slice thickness of 1.5 mm and an in-plane spatial resolution of 0.98 × 0.98–1.17 × 1.17 mm2. After an 8-h fast, patients were injected with 370–666 MBq (10–18 mCi) of 18F-FDG, with imaging taking place 60 min later. PET data were acquired in 2-dimensional mode, with a 3- to 5-min acquisition time per bed position. The PET images were then reconstructed with an ordered-subset expectation maximization algorithm, using the CT data for attenuation correction. The spatial resolution of the original PET image was 2.34 × 2.34 × 3.27 mm3.
Image Preprocessing
The primary tumor and involved lymph nodes were separately contoured on both pre- and midtreatment CT scans for radiation treatment planning or replanning purposes by an attending radiation oncologist specializing in head and neck cancer. These contours were computationally extracted using in-house–developed software in MATLAB (The MathWorks) for subsequent analyses. We then spatially aligned the pre- and midtreatment contrast-enhanced CT images and the PET and CT images using the rigid registration function of the well-validated Elastix software (21). The use of rigid rather than deformable registration is necessary to detect soft-tissue changes and tumor response. Moreover, the quality of the registration was carefully inspected for alignment of the bony structures, including the skull, mandible, and cervical vertebrae. Each CT slice was manually inspected by a radiation oncologist for metal artifacts due to dental fillings. Those slices with severe artifacts were excluded in subsequent analysis, and the effect on proposed features was adjusted for and evaluated in detail. To reduce the variability in PET SUV, we normalized the SUV by the average background activity in a circular region of interest in the aortic arch (22). All images were resampled at an isotropic spatial resolution of 1.0 mm3 via the MATLAB (R2017b) function imresize3.
Identification and Validation of Habitat Regions by Integrated PET and CT Analysis
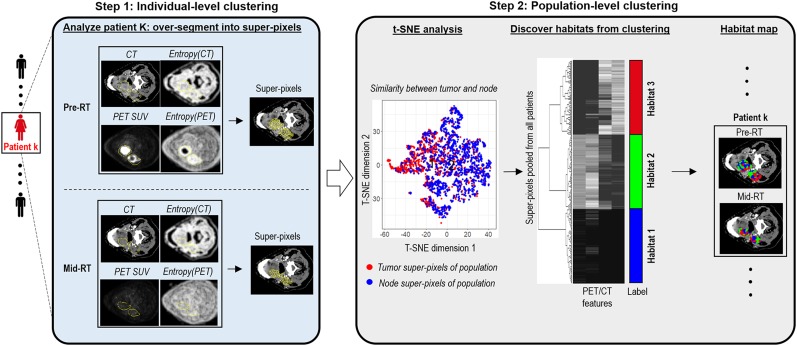
We proposed a robust partitioning method to identify subregions within the primary tumor and involved lymph nodes. The method consists of a 2-step clustering process as outlined in Figure 2. First, at the patient level, the primary tumor and involved nodes on pre- and midtreatment images were oversegmented into numerous superpixels, respectively. In detail, given precalculated 4-feature maps, including PET SUV, CT number, entropy of PET SUV, and entropy of CT number, the SLIC algorithm (23) was used to generate superpixels, with squared Euclidean distance as the similarity metric. We implemented this with the MATLAB (R2017b) function superpixels3. Each superpixel contains connected voxels with similar imaging characteristics as defined by CT number, PET SUV, and spatial heterogeneity measured by local entropy maps. The similarity between superpixels in the primary tumor and nodes was visualized with t-SNE (24,25).
FIGURE 2.
Proposed habitat discovery framework. Method consists of 2-stage clustering process: individual-level clustering based on CT number, PET SUV, and their local entropy maps, followed by population-level consensus clustering. pre-RT = pretreatment of radiation therapy; mid-RT = midtreatment of radiation therapy; t-SNE = t-distributed stochastic neighbor embedding.
Next, we aggregated all patients’ superpixels in tumor and nodes at pre- and midtreatment images and performed consensus clustering at the population level to uncover the number and membership of habitat regions, whereby superpixels with similar image phenotypes were merged to form a habitat. In detail, each superpixel was characterized by these 4 parameters averaged over embraced voxels. Cumulating superpixels for all patients in a global matrix, consensus clustering (26) was applied to explore inter- and intrapatient similarity and discover consistent patterns in the population. Here, we used the partition-around-medoids clustering algorithm and Euclidean distance. We performed 10,000 bootstraps with 80% resampling of the superpixels. The candidate cluster number was tested from 2 to 10, and the optimal one was selected to produce the most stable consensus matrix and the least ambiguous cluster assignments across permuted clustering runs. In particular, the optimal cluster number corresponded to the largest cluster number that induced the minimum incremental change in the area under the cumulative distribution function curves, while maximizing consensus within clusters and minimizing the ambiguity rate in cluster assignments. We implemented it with R package ConsensusClusterPlus. We independently applied the proposed pipeline to discover habitats in the training and validation cohorts, and quantitatively assessed the consistency of the habitats across two cohorts using the in-group proportion statistic.
Quantitative Spatiotemporal Habitat Evolution-Based Image Features
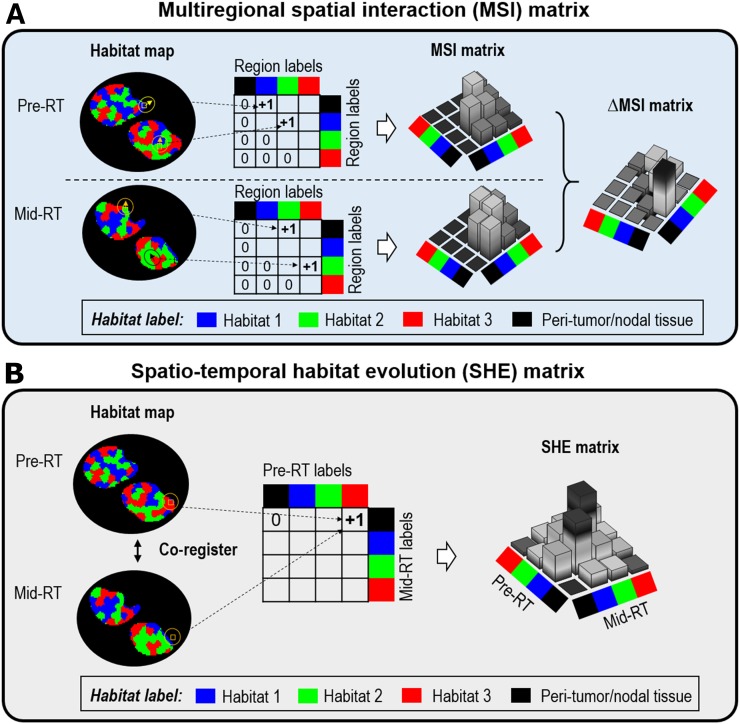
To characterize the complex response patterns during treatment, we proposed novel quantitative spatiotemporal image features based on the habitats identified earlier. Specifically, 3 types of image features were investigated, detailed in Table 2 and Supplemental Table 1. First, we computed the volumetric change in individual habitat regions from pre- to midtreatment (i.e., ∆Volhabitat). Second, we investigated how the overall spatial arrangement of habitat regions evolved during treatment, by comparing the multiregional spatial interaction (MSI) matrix between pre- and midtreatment images (Fig. 3A). Briefly, the MSI matrix summarized the spatial cooccurrence statistics among different habitat regions in the same image. The response metric was calculated as the difference in MSI between 2 time points (i.e., ∆MSI = MSImid − MSIpre).
TABLE 2.
Details of Investigated Imaging Features from PET and CT, Including 27 Habitat-Related Features and 13 Conventional Imaging Markers
| Type | Details | n |
| Habitat-related features | ||
| Change of individual habitat region’s burden | ∆Volhabitat i where i = 1,2,3 | 3 |
| Change for connected habitat region burden measured by MSI matrix | ∆MSI i, where i = 1,…,9 | 9 |
| Habitat progression via spatiotemporal habitat evolution (SHE) matrix | SHE i, where i = 1,…,15 | 15 |
| Conventional features | ||
| Tumor and node burden from CT | (a) Pre.Vol: volume from pre-RT CT; (b) Mid.Vol: volume from mid-RT CT; (c) ∆VolCT: volume change | 3 |
| Metabolic activity of tumor and node regions from PET | Pre-RT PET: (a) Pre.SUVmax; (b) Pre.MTV2.5; (c) Pre.MTVFCM | 9 |
| Mid-RT PET: (d) Mid.SUVmax; (e) Mid.MTV2.5; (f) Mid.MTVFCM | ||
| Change: (g) ∆SUVmax; (h) ∆MTV2.5; (i) ∆MTVFCM | ||
| Radiomics signature of tumor | Radiomics: prediction model built from set of 600 radiomics features (321 from CT, 279 from PET SUV) | 1 |
∆ = mid − pre; pre-RT = pretreatment radiotherapy; mid-RT = midtreatment radiotherapy; MTVFCM = MTV adaptively defined using fuzzy c-means algorithm; MTV2.5 = MTV defined using SUV of 2.5 as cutoff.
FIGURE 3.
(A) Implementation of MSI matrix, which summarizes cooccurrence statistics among different habitat regions and is computed separately for pre- and midtreatment image. ∆MSI (MSImidtreatment − MSIpretreatment) is calculated to quantify change of spatial habitat interaction. (B) Implementation of spatiotemporal habitat evolution matrix to summarize temporal cooccurrence statistics among spatially coregistered habitat maps, where voxel-level evolution of habitats from baseline to midtreatment is characterized. In both cases, we explicitly included surrounding tissue as distinct habitat region to account for its relationship with tumor. pre-RT = pretreatment of radiation therapy; mid-RT = midtreatment of radiation therapy.
The third category of image features aimed to characterize the response pattern of the habitats at the voxel level (Fig. 3B). We proposed a spatiotemporal habitat evolution matrix, which summarized the temporal cooccurrence statistics among different habitats between the baseline and midtreatment images. To compute the spatiotemporal habitat evolution matrix, we compared the habitat labels at corresponding voxels between coregistered pre- and midtreatment habitat maps, and we computed the counts among different habitats by iterating through all voxels. For both MSI and spatiotemporal habitat evolution matrices, we explicitly included the peritumoral tissue as a distinct habitat region to account for its relationship with the tumor and node.
Evaluation for Reproducibility of the Habitat-Related Image Features
We assessed the stability of the proposed image features against variations in tumor delineation and image registration, as well as the effect of dental artifacts. First, the primary tumor and involved lymph nodes were independently recontoured by a second radiation oncologist for patients in the training cohort. We repeated the whole computational analysis based on the new contours and investigated the feature reproducibility. Second, to assess the impact of image registration on features based on the spatiotemporal habitat evolution matrix, we repeated the registration with a different set of parameters in Elastix and compared the resultant features. Third, we assessed the impact of dental artifacts on the proposed features by randomly excluding a similar number of CT slices in patients without dental artifacts. Because certain image slices are excluded in some patients, this step will result in smaller habitat volumes, leading to a potential bias in the computed features. To compensate for this bias, we adjusted for the computed features with a multiplicative factor assuming similar habitat distributions in the excluded slices. We compared the adjusted image features with the ground truth computed using the complete scan. The intraclass correlation coefficient was used to evaluate the reproducibility of the proposed image features.
Training and Validation of Imaging Signature for Predicting Progression-Free Survival (PFS)
On the basis of the proposed habitat-based image features, we fitted a multivariable Cox proportional-hazards regression model in the training cohort to predict PFS. This step was combined with the least-absolute-shrinkage-and-selection-operator algorithm to mitigate the risk of overfitting. Before model construction, we assessed the pairwise correlation between features and removed redundant features with a Pearson correlation coefficient of more than 0.9 and lower predictive power in terms of the C-index to avoid colinearity in fitting the Cox model. Tenfold cross-validation was applied and repeated 100 times to minimize potential selection bias, and the most frequently selected features (>90%) were used to refit the final Cox model.
Then, we evaluated the performance of the proposed imaging signature for predicting PFS in the independent validation cohort. The ability to predict 2-year PFS (a clinically relevant endpoint in oropharyngeal cancer) was assessed by using survival receiver operating characteristic curve. We also compared the prediction accuracy for the habitat-based imaging signature with conventional imaging metrics and the combined model. In particular, conventional imaging metrics contain pretreatment, midtreatment, and change of in volume (at CT), SUVmax, and MTV2.5 measured from both tumor and node regions. In addition, since MTV2.5 may be sensitive to small and heterogeneous lesions (27), we adopted a previous validated study (28) to implement a fuzzy c-means algorithm to adaptively define MTV (MTVFCM). Moreover, we have trained radiomics model based on a set of 600 features extracted from PET and CT via pyradiomics (29) (Supplemental Table 1), as well as a clinicopathologic model based on T stage, N stage, smoking status, and HPV.
We further evaluated the habitat-based model’s performance to stratify patients within subgroups defined by HPV status, smoking history, and stage. Finally, we performed multivariable Cox analysis separately in the training and validation cohorts and tested whether the proposed imaging signature provided independent prognostic value beyond clinical and pathologic risk factors (2), including baseline tumor volume, T stage, N stage, HPV status, smoking history, and re-planning status at mid-treatment.
Statistical Analysis
The concordance index (c-index) or the Harrell C statistic was used to assess prognostic accuracy. Kaplan–Meier analysis and log-rank testing were used to evaluate patient stratification into different risk groups. The cutoff was based on the median risk score in the training cohort to divide the patients into high- and low-risk groups, and the same cutoff was applied to the validation cohort. The hazard ratio was used to measure the degree of survival difference, and the 95% confidence interval was reported. All statistical tests were 2-sided, with a P value of less than 0.05 considered significant. All statistical analyses were performed in R (R Foundation for Statistical Computing).
RESULTS
Three Habitat Regions Associated with Distinct Imaging Phenotypes
We first assessed the similarity in PET and CT imaging phenotypes between superpixels in the primary tumor and involved lymph nodes. As shown in t-distributed stochastic neighbor embedding plots in Supplemental Figures 2 and 3, superpixels extracted from tumor and node regions at pre- or midtreatment images were well mixed for both the training and validation cohorts, confirming our hypothesis. We therefore merged superpixels in the primary tumor and involved nodes in subsequent analyses.
We independently applied our proposed habitat discovery pipeline for the training and validation cohorts and identified 3 distinct habitats that were highly consistent between the 2 cohorts (Supplemental Figs. 4A and 5A), with in-group proportions of 0.97, 0.99, and 0.99 (P < 0.001) for habitats 1, 2, and 3, respectively. The optimal number of habitats was further confirmed by the area under cumulative-distribution-function curves in Supplemental Figures 4B and 5B.
On the basis of the distributions of the 4 imaging features in 3 habitats (Supplemental Figs. 6 and 7), we observed that habitat 3 was associated with the highest PET SUV and the highest PET entropy, suggesting it may contain the most metabolically active and heterogeneous component. On the other hand, habitat 2 was associated with the highest CT number and CT entropy, suggesting it may contain the most contrast-enhanced and heterogeneous component. By contrast, habitat 1 was associated with the lowest value for all 4 imaging features, suggesting it may contain the unenhanced, metabolically inactive, and homogeneous component. These results were consistent between the training and validation cohorts. The distribution of superpixels across 3 habitats is similar between the tumor and node (Supplemental Fig. 8). Moreover, the change from pre- to midtreatment also follows the same trend, with an increase in habitats 1 and 2 and a decrease in habitat 3.
Habitat Image Features and Stability with Respect to Technical Factors
We applied the proposed computational pipeline to extract a total of 27 habitat-based image features (Table 2; Supplemental Table 1). On the basis of the pairwise correlation map of proposed image features (Supplemental Fig. 9), we removed 3 redundant and less prognostic features before fitting the Cox model, including ∆Volhabitat1, ∆Volhabitat2, and ∆Volhabitat3. To assess the effect of tumor delineation on the proposed features, we generated 2 independent sets of contours and calculated the extent of overlap by dice coefficients (Supplemental Fig. 10). Most of the proposed image features showed good reproducibility, with an average intraclass correlation coefficient of 0.82 (Supplemental Fig. 11). Similarly, the image features derived from different image registration settings were also highly reproducible, with an average intraclass correlation coefficient of 0.95 (Supplemental Fig. 12). Finally, although around two thirds of patients had some dental artifacts, these affected only a small portion of the target volume: 12% for the primary tumor and 3.4% for the lymph nodes on average. By randomly excluding slices for patients with otherwise complete images, we observed that the features were highly consistent with those computed from the complete image (average intraclass correlation coefficient, 0.90; Supplemental Fig. 13). These results demonstrate the robustness of the proposed image features against variations in contouring and registration, as well as dental artifacts.
Better Prediction of PFS with Habitat-Based Imaging Signature Than with Conventional Imaging Metrics
On the basis of the refined image feature set, we built a predictive model for PFS using the training cohort. As shown in Supplemental Table 2, the final imaging signature contained 4 features, including a volumetric change in interactions between habitat 1 and peritumoral tissue, as well as between habitats 1 and 3, the volume of spatially persistent habitat 3, and the volume of peritumoral tissue at baseline evolving to habitat 2 at midtreatment. The imaging signature did not correlate with any of the conventional image features (Pearson correlation < 0.5, Supplemental Fig. 14).
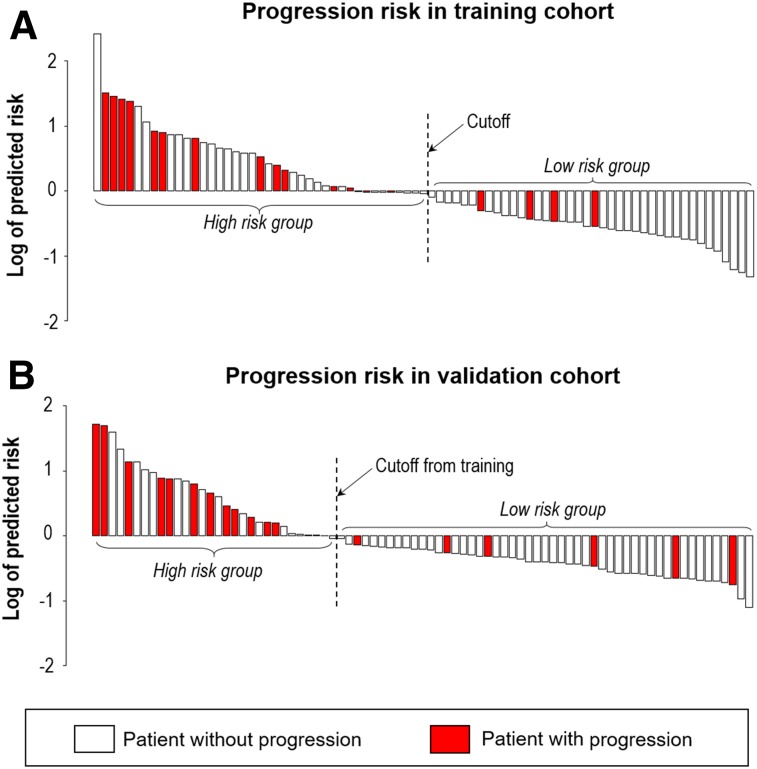
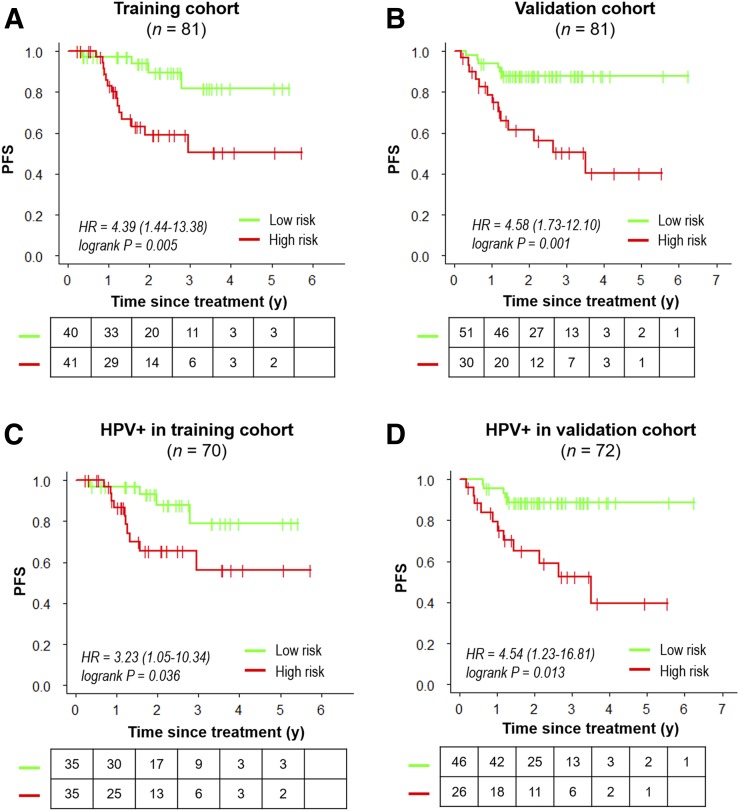
With the median as the cutoff (Fig. 4), the imaging signature separated patients in the training cohort into 2 groups with a significantly different PFS (hazard ratio, 4.39; 95% confidence interval, 1.44–13.4; log-rank P = 0.005; Fig. 5A). At the same cutoff, the imaging signature also stratified patients for PFS in the validation cohort (hazard ratio, 4.58; 95% confidence interval, 1.73–12.1; log-rank P = 0.001; Fig. 5B). 2-y PFS rates for high-risk and low-risk groups were 59.1% versus 89.4% in training cohort and 61.4% versus 87.8% in the validation cohort. Importantly, this stratification remained significant in both training and validation cohorts within the subgroup of HPV-positive tumors (Figs. 5C and 5D), patients with at least 10 pack-years of smoking (Supplemental Figs. 15A and 15B), patients with stage IV disease (Supplemental Figs. 15C and 15D), and patients treated at different time frames (Supplemental Figs. 15E and 15F).
FIGURE 4.
Waterfall plot of predicted risk with proposed habitat evolution-based imaging signature for training cohort (A) and validation cohort (B).
FIGURE 5.
Kaplan–Meier curves of progression-free survival (PFS), where patients are stratified by median risk score according to proposed habitat evolution-based imaging signature in training cohort. Plots are for overall training cohort (A), overall validation cohort (B), HPV-positive subgroup in training (C), and HPV-positive subgroup in validation (D). HR = hazard ratio.
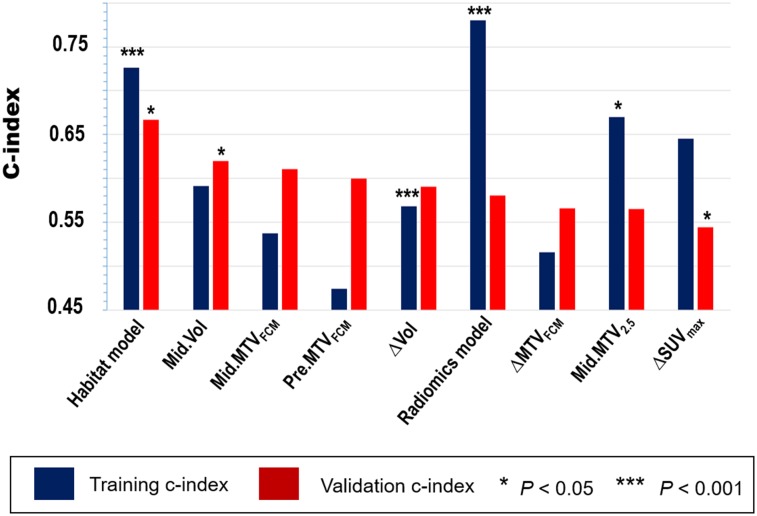
We then compared the prediction performance of the proposed imaging signature with 12 conventional imaging features and the radiomics signature (Supplemental Tables 1 and 3). As shown in Figure 6 and Supplemental Figure 16, the proposed imaging signature outperformed all conventional features in the validation cohort, including midtreatment CT-based tumor volume or PET-based MTV (C-index, 0.66 vs. 0.61–0.62). Classification analysis of predicting 2-y PFS showed similar results (Supplemental Fig. 17). Despite a superior performance in the training cohort, the radiomics signature failed to predict PFS in the validation cohort (C-index, 0.58; P = 0.467). Combining conventional imaging features with the habitat-based imaging signature did not improve the prediction performance, with a C-index of 0.64 (P = 0.875) in the validation cohort. In multivariable analysis, the proposed imaging signature remained an independent prognostic factor after adjusting for clinicopathologic variables and baseline tumor volume (Table 3).
FIGURE 6.
Accuracy of predicting PFS as measured by C-index for habitat-based imaging signature, compared with top-ranked conventional imaging features from PET and CT images extracted, respectively, at pretreatment, midtreatment, or change (mid–pre). comparison was performed separately in training and validation cohorts. features were ranked by C-index of validation. ∆ = mid − pre; MTVFCM = MTV adaptively defined using fuzzy c-means algorithm; MTV2.5 = MTV defined using SUV of 2.5 as cutoff.
TABLE 3.
Univariate and Multivariable Analyses of Proposed Imaging Signature and Clinicopathologic Factors for Predicting PFS
| Training cohort |
Validation cohort |
|||||||||||
| Univariate |
Multivariate |
Univariate |
Multivariate |
|||||||||
| Predictors | HR | 95% CI | P | HR | 95% CI | P | HR | 95% CI | P | HR | 95% CI | P |
| Habitat imaging signature (continuous) | 1.36 | 1.10–1.68 | 0.004 | 1.30 | 1.02–1.66 | 0.028 | 1.37 | 1.05–1.79 | 0.021 | 1.41 | 1.01–1.98 | 0.048 |
| T* | 2.07 | 0.78–5.53 | 0.145 | — | — | 0.866 | 1.99 | 0.78–5.07 | 0.147 | — | — | 0.650 |
| N† | 3.98 | 0.53–29.93 | 0.180 | — | — | 0.258 | 2.05 | 0.83–5.06 | 0.117 | — | — | 0.405 |
| HPV‡ | 0.50 | 0.17–1.53 | 0.225 | — | — | 0.197 | 0.56 | 0.16–1.93 | 0.359 | — | — | 0.867 |
| Smoking history§ | 1.03 | 0.38–2.75 | 0.957 | — | — | 0.675 | 1.07 | 0.42–2.72 | 0.894 | — | — | 0.623 |
| Baseline gross tumor volume at CT | 1.14 | 0.74–1.74 | 0.561 | — | — | 0.757 | 1.52 | 1.08–2.12 | 0.015 | 1.70 | 1.08–2.69 | 0.023 |
| Adaptive planning at midtreatment‖ | 2.54 | 0.84–7.75 | 0.100 | — | — | 0.119 | 1.14 | 0.33–3.91 | 0.887 | — | — | 0.445 |
T1–3 as 0, T4 as 1.
N0–N2a as 0, N2b–N3 as 1.
‡HPV+ as 1, HPV− as 0.
Smoking ≤10 pack-years as 0, otherwise as 1.
Plan change as 1, no change as 0.
HR = hazard ratio; CI = confidence interval.
DISCUSSION
We developed and validated a novel habitat evolution-based imaging signature to assess early response and predict treatment outcomes of OPSCC. The imaging signature provided independent prognostic information beyond established clinical factors and further stratified patients into different risk groups within HPV-positive OPSCC. The imaging signature has the potential to refine the selection of low-risk HPV-positive patients who are eligible for treatment deintensification. A commonly accepted criterion for eligibility is that these patients should have a 2-y PFS above 90%. Currently, our model stratified the low-risk HPV-positive patients who had 2-y PFS of 88.6%. This is a promising result but requires further validation. On the other hand, the prognosis of HPV-negative disease remains poor with concurrent chemoradiotherapy, and novel treatment strategies such as immunotherapy (30) may be needed for these patients. The proposed imaging signature could potentially be used to identify high-risk patients who might benefit from adjuvant immunotherapy.
To our knowledge, this is the largest study to investigate the prognostic value of midtreatment 18F-FDG PET and CT imaging in OPSCC. Previously, several studies have investigated imaging characteristics associated with treatment outcomes in OPSCC (9,11,12,16). Most of these studies are based on either pretreatment or posttreatment imaging, however. By contrast, midtreatment imaging has the unique advantage of allowing assessment of early response, which may be better correlated with disease control and outcomes, and at the same time still offers the opportunity to modify treatment. In a recent study, the change in primary tumor volume measured at midtreatment CT compared with baseline was shown to be associated with locoregional recurrence in OPSCC (20). In a previous study of a smaller cohort of 74 patients, among traditional imaging metrics midtreatment MTV was found to be the most important one for predicting outcomes (11). Our current study with 162 patients confirmed these findings, and the new habitat imaging signature further improved the prediction accuracy over midtreatment tumor volume at CT and metabolic tumor volume at PET.
One key aspect of our study is that we analyzed both the primary tumor and the involved lymph nodes, whereas previous studies have focused mostly on the primary tumor only. Patients with OPSCC, especially HPV-positive disease, tend to have early development of regional metastases to the lymph nodes. Therefore, incorporating involved nodal disease into the analysis can provide a more complete evaluation and direct reflection of the metastatic potential. Indeed, several studies have recently shown that specific imaging characteristics of the involved nodes could play a more important role in predicting outcomes than features derived from the primary tumor. Our current study is consistent with these recent findings and suggests that further investigation of the imaging characteristics of nodal disease for predicting response or prognosis may be warranted. Here, we combined the habitat features extracted from primary tumor and nodes to mitigate the overfitting risk with limited samples. In the future, it is worth exploring features of these 2 regions separately in a larger cohort.
Multiregion sequencing studies have revealed significant variations in the genetic makeup and molecular pathways across different regions in the same tumor (31). Image-based partitioning could be used to identify aggressive subregions that are important for determining prognosis and treatment response (32–34). Gillies et al. used a threshold-based method to segment intratumoral regions (i.e., habitats) with prognostic value in glioblastoma (35). Cui et al. showed that radiomic analysis of intratumoral subregions predicted survival better than whole-tumor imaging metrics in glioblastoma (36). Spatial heterogeneity among tumor habitats has also been explored in breast cancer to predict chemotherapy response and disease recurrence (37,38). Built on these pilot studies, we further extended the habitat imaging approach to investigate primary tumor and nodal regions at baseline and midtreatment imaging. The proposed novel spatiotemporal habitat evolution-based image features serve to characterize the complex response patterns and predict prognosis in OPSCC. We envision that the proposed approach will be applicable to other cancer types to dissect intratumor heterogeneity and track tumor evolution through longitudinal imaging analysis.
Our study differs from the traditional high-throughput radiomics approaches that extract hundreds of computational image features and are usually performed on the primary tumor at baseline CT. Recently, concerns have been raised regarding the stability, reproducibility, and interpretability of high-throughput radiomic features and signatures (39,40). By contrast, we proposed volumetric measurement of intratumoral/nodal subregions (habitats) as image features, as they are well-defined physical quantities, are simple to interpret, and may be biologically relevant. In addition, they may be less dependent on technical variations in image acquisition than traditional intensity or texture-based radiomic features. The prognostic accuracy of the habitat imaging signature is robust to different splits of training and validation sets. Also, the results are highly reproducible when we split the patients by their treatment time, suggesting robustness of proposed analysis pipeline.
One limitation of the study is that it reflects the experience at a single institution and is based on retrospective data. The proposed imaging signature should be further tested in independent, prospective studies. Issues such as repeatability and reproducibility should be addressed for multicenter validation as proposed in previous studies (41–43). Another limitation is that the cohort includes both HPV-positive and HPV-negative tumors. When we controlled for HPV status, the imaging signature remained independently prognostic in multivariable analysis and also stratified patients with HPV-positive tumors. Future work should analyze patients separately according to HPV status given the distinct biology between the 2 diseases. In the future, it may be interesting to explore deep-learning approaches, in particular, convolutional neural networks, which can automate the extraction of image features with prognostic potential, but this will require a large number of patients for training purposes.
CONCLUSION
We proposed novel habitat evolution-based image features to assess early response. We developed and independently validated an imaging signature to predict treatment outcomes, and this imaging signature improved on traditional imaging response metrics. If prospectively validated, the proposed imaging signature could potentially be used to refine patient selection for individualized risk-adaptive therapy in oropharyngeal cancer.
DISCLOSURE
This research was partially supported by National Institutes of Health grants R01 CA222512, R01 CA193730, and K99 CA218667. No other potential conflict of interest relevant to this article was reported.
KEY POINTS
QUESTION: Can parameters defined at baseline and midtreatment 18F-FDG PET/CT imaging predict disease relapse in oropharyngeal cancer?
PERTINENT FINDINGS: In a cohort of 162 patients with oropharyngeal cancer treated with concurrent chemoradiotherapy, a novel habitat evolution-based imaging signature was developed and independently validated to assess early response and predict treatment outcomes. The imaging signature provided independent prognostic information beyond established clinicopathologic factors and further improved on traditional imaging response metrics.
IMPLICATIONS FOR PATIENT CARE: Serial 18F-FDG PET/CT imaging may be used to identify patients at high risk of disease relapse and guide risk-adaptive therapy in oropharyngeal cancer.
Supplementary Material
REFERENCES
- 1.Chaturvedi AK, Engels EA, Pfeiffer RM, et al. Human papillomavirus and rising oropharyngeal cancer incidence in the United States. J Clin Oncol. 2011;29:4294–4301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ang KK, Harris J, Wheeler R, et al. Human papillomavirus and survival of patients with oropharyngeal cancer. N Engl J Med. 2010;363:24–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen AM, Felix C, Wang PC, et al. Reduced-dose radiotherapy for human papillomavirus-associated squamous-cell carcinoma of the oropharynx: a single-arm, phase 2 study. Lancet Oncol. 2017;18:803–811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Marur S, Li S, Cmelak AJ, et al. E1308: phase II trial of induction chemotherapy followed by reduced-dose radiation and weekly cetuximab in patients with HPV-associated resectable squamous cell carcinoma of the oropharynx—ECOG-ACRIN Cancer Research Group. J Clin Oncol. 2017;35:490–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gillison ML, Trotti AM, Harris J, et al. Radiotherapy plus cetuximab or cisplatin in human papillomavirus-positive oropharyngeal cancer (NRG Oncology RTOG 1016): a randomised, multicentre, non-inferiority trial. Lancet. 2019;393:40–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.O’Sullivan B, Huang SH, Su J, et al. Development and validation of a staging system for HPV-related oropharyngeal cancer by the International Collaboration on Oropharyngeal cancer Network for Staging (ICON-S): a multicentre cohort study. Lancet Oncol. 2016;17:440–451. [DOI] [PubMed] [Google Scholar]
- 7.Rohde M, Nielsen AL, Pareek M, et al. PET/CT versus standard imaging for prediction of survival in patients with recurrent head and neck squamous cell carcinoma. J Nucl Med. 2019;60:592–599. [DOI] [PubMed] [Google Scholar]
- 8.Aerts HJ, Velazquez ER, Leijenaar RT, et al. Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nat Commun. 2014;5:4006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Leijenaar RT, Carvalho S, Hoebers FJ, et al. External validation of a prognostic CT-based radiomic signature in oropharyngeal squamous cell carcinoma. Acta Oncol. 2015;54:1423–1429. [DOI] [PubMed] [Google Scholar]
- 10.Vallières M, Kay-Rivest E, Perrin LJ, et al. Radiomics strategies for risk assessment of tumour failure in head-and-neck cancer. Sci Rep. 2017;7:10117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pollom EL, Song J, Durkee BY, et al. Prognostic value of midtreatment FDG-PET in oropharyngeal cancer. Head Neck. 2016;38:1472–1478. [DOI] [PubMed] [Google Scholar]
- 12.M.D. Anderson Cancer Center Head and Neck Quantitative Imaging Working Group. Investigation of radiomic signatures for local recurrence using primary tumor texture analysis in oropharyngeal head and neck cancer patients. Sci Rep. 2018;8:1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Desseroit MC, Tixier F, Weber WA, et al. Reliability of PET/CT shape and heterogeneity features in functional and morphologic components of non-small cell lung cancer tumors: a repeatability analysis in a prospective multicenter cohort. J Nucl Med. 2017;58:406–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wong KH, Panek R, Welsh L, et al. The predictive value of early assessment after 1 cycle of induction chemotherapy with 18F-FDG PET/CT and diffusion-weighted MRI for response to radical chemoradiotherapy in head and neck squamous cell carcinoma. J Nucl Med. 2016;57:1843–1850. [DOI] [PubMed] [Google Scholar]
- 15.Welch ML, McIntosh C, Haibe-Kains B, et al. Vulnerabilities of radiomic signature development: the need for safeguards. Radiother Oncol. 2019;130:2–9. [DOI] [PubMed] [Google Scholar]
- 16.Kwan JYY, Su J, Huang SH, et al. Radiomic biomarkers to refine risk models for distant metastasis in HPV-related oropharyngeal carcinoma. Int J Radiat Oncol Biol Phys. 2018;102:1107–1116. [DOI] [PubMed] [Google Scholar]
- 17.Leger S, Zwanenburg A, Pilz K, et al. CT imaging during treatment improves radiomic models for patients with locally advanced head and neck cancer. Radiother Oncol. 2019;130:10–17. [DOI] [PubMed] [Google Scholar]
- 18.Bogowicz M, Riesterer O, Ikenberg K, et al. Computed tomography radiomics predicts HPV status and local tumor control after definitive radiochemotherapy in head and neck squamous cell carcinoma. Int J Radiat Oncol Biol Phys. 2017;99:921–928. [DOI] [PubMed] [Google Scholar]
- 19.Wu J, Gensheimer MF, Zhang N, et al. Integrating tumor and nodal imaging characteristics at baseline and mid-treatment computed tomography scans to predict distant metastasis in oropharyngeal cancer treated with concurrent chemoradiotherapy. Int J Radiat Oncol Biol Phys. 2019;104:942–952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kabarriti R, Brodin NP, Lundgren G, et al. Early response assessment on mid-treatment computed tomography predicts for locoregional recurrence in oropharyngeal cancer patients treated with definitive radiation therapy. Int J Radiat Oncol Biol Phys. 2018;102:1036–1045. [DOI] [PubMed] [Google Scholar]
- 21.Klein S, Staring M, Murphy K, Viergever MA, Pluim JP. Elastix: a toolbox for intensity-based medical image registration. IEEE Trans Med Imaging. 2010;29:196–205. [DOI] [PubMed] [Google Scholar]
- 22.Bauer C, Sun S, Sun W, et al. Automated measurement of uptake in cerebellum, liver, and aortic arch in full‐body FDG PET/CT scans. Med Phys. 2012;39:3112–3123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Achanta R, Shaji A, Smith K, Lucchi A, Fua P, Süsstrunk S. SLIC superpixels compared to state-of-the-art superpixel methods. IEEE Trans Pattern Anal Mach Intell. 2012;34:2274–2282. [DOI] [PubMed] [Google Scholar]
- 24.van der Matten L, Hinton G. Visualizing data using t-SNE. J Mach Learn Res. 2008;9:2579–2605. [Google Scholar]
- 25.Raidou RG, van der Heide UA, Dinh CV, et al. Visual analytics for the exploration of tumor tissue characterization. Comput Graph Forum. 2015;34:11–20. [Google Scholar]
- 26.Monti S, Tamayo P, Mesirov J, Golub T. Consensus clustering: a resampling-based method for class discovery and visualization of gene expression microarray data. Machine Learning. 2003;52:91–118. [Google Scholar]
- 27.Zaidi H, El Naqa I. PET-guided delineation of radiation therapy treatment volumes: a survey of image segmentation techniques. Eur J Nucl Med Mol Imaging. 2010;37:2165–2187. [DOI] [PubMed] [Google Scholar]
- 28.Belhassen S, Zaidi H. A novel fuzzy C‐means algorithm for unsupervised heterogeneous tumor quantification in PET. Med Phys. 2010;37:1309–1324. [DOI] [PubMed] [Google Scholar]
- 29.Van Griethuysen JJ, Fedorov A, Parmar C, et al. Computational radiomics system to decode the radiographic phenotype. Cancer Res. 2017;77:e104–e107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ferris RL, Blumenschein G, Jr, Fayette J, et al. Nivolumab for recurrent squamous-cell carcinoma of the head and neck. N Engl J Med. 2016;375:1856–1867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gerlinger M, Rowan AJ, Horswell S, et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med. 2012;366:883–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gatenby RA, Grove O, Gillies RJ. Quantitative imaging in cancer evolution and ecology. Radiology. 2013;269:8–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wu J, Gensheimer MF, Dong X, et al. Robust intratumor partitioning to identify high-risk subregions in lung cancer: a pilot study. Int J Radiat Oncol Biol Phys. 2016;95:1504–1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Even AJG, Reymen B, La Fontaine MD, et al. Clustering of multi-parametric functional imaging to identify high-risk subvolumes in non-small cell lung cancer. Radiother Oncol. 2017;125:379–384. [DOI] [PubMed] [Google Scholar]
- 35.Zhou M, Chaudhury B, Hall LO, Goldgof DB, Gillies RJ, Gatenby RA. Identifying spatial imaging biomarkers of glioblastoma multiforme for survival group prediction. J Magn Reson Imaging. 2017;46:115–123. [DOI] [PubMed] [Google Scholar]
- 36.Cui Y, Tha KK, Terasaka S, et al. Prognostic imaging biomarkers in glioblastoma: development and independent validation on the basis of multiregion and quantitative analysis of MR images. Radiology. 2016;278:546–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wu J, Gong G, Cui Y, Li R. Intratumor partitioning and texture analysis of dynamic contrast-enhanced (DCE)-MRI identifies relevant tumor subregions to predict pathological response of breast cancer to neoadjuvant chemotherapy. J Magn Reson Imaging. 2016;44:1107–1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wu J, Cao G, Sun X, et al. Intratumoral spatial heterogeneity at perfusion MR imaging predicts recurrence-free survival in locally advanced breast cancer treated with neoadjuvant chemotherapy. Radiology. 2018;288:26–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Berenguer R, Pastor-Juan MDR, Canales-Vázquez J, et al. Radiomics of CT features may be nonreproducible and redundant: influence of CT acquisition parameters. Radiology. 2018;288:407–415. [DOI] [PubMed] [Google Scholar]
- 40.Vallières M, Zwanenburg A, Badic B, Cheze Le Rest C, Visvikis D, Hatt M. Responsible radiomics research for faster clinical translation. J Nucl Med. 2018;59:189–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Orlhac F, Boughdad S, Philippe C, et al. A postreconstruction harmonization method for multicenter radiomic studies in PET. J Nucl Med. 2018;59:1321–1328. [DOI] [PubMed] [Google Scholar]
- 42.Press RH, Shu HG, Shim H, et al. The use of quantitative imaging in radiation oncology: a quantitative imaging network (QIN) perspective. Int J Radiat Oncol Biol Phys. 2018;102:1219–1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kurland BF, Peterson LM, Shields AT, et al. Test-retest reproducibility of 18F-FDG PET/CT uptake in cancer patients within a qualified and calibrated local network. J Nucl Med. 2019;60:608–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.