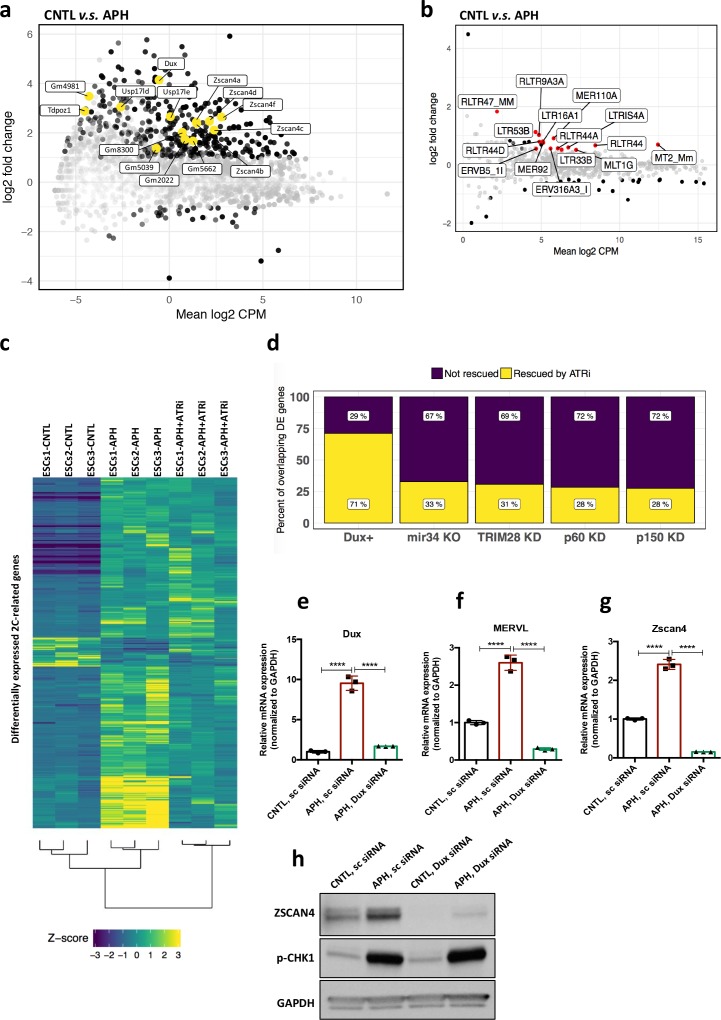
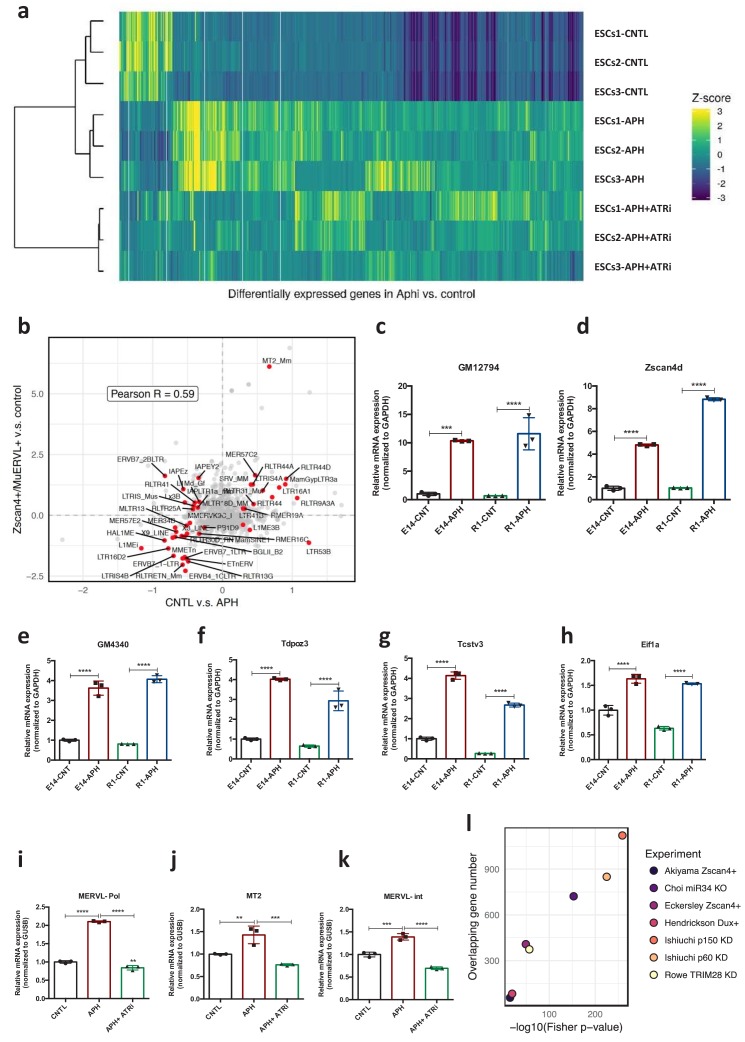
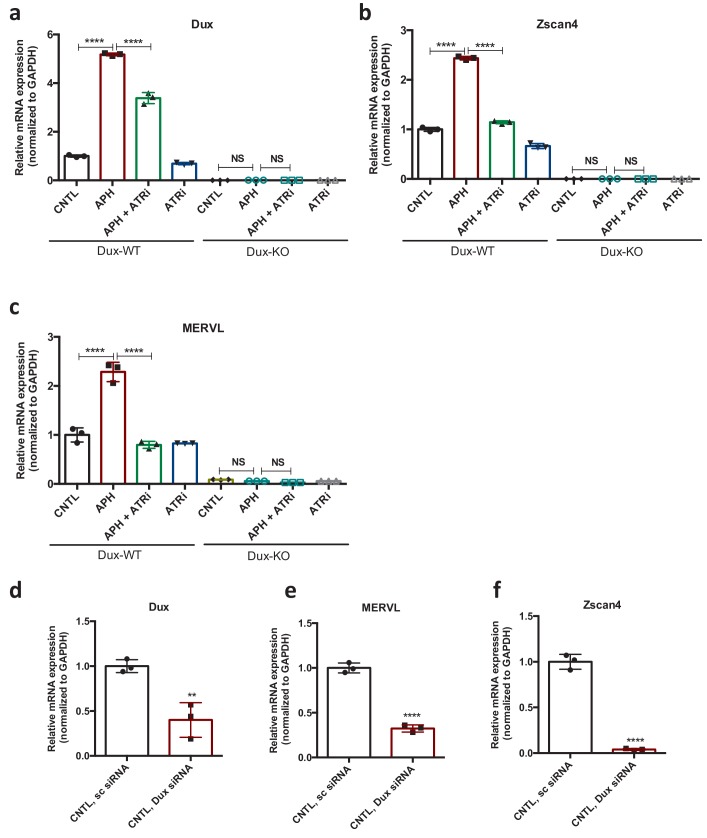
Figure 3. ATR induces transcriptional signature of 2C-like cells in ESCs.
(a) MA plot showing gene expression in ESCs treated with APH in comparison to control. Key 2C-specific genes are highlighted (b) MA plot showing retrotransposons expression upon APH treatment. (c) Heatmap showing the robust z-scores for 2C-specific genes in the indicated samples. 2C-related genes were identified by performing a differential expression analysis on ZSCAN4+/MERVL+v.s. ZSCAN4-/ MERVL- ESCs from Eckersley-Maslin et al. (2016). (d) Bar plot displaying the percentage of ATR-dependent differentially expressed genes among the ones shared between APH-treated ESCs and each dataset. (e–g) RT-qPCR analysis of Dux KD ESCs for Dux and Zscan4d genes, and MERVL upon treatment with APH. (h) Immunoblot for p-CHK1 and ZSCAN4 proteins upon treatment with APH in Dux KD ESCs in comparison with control ESCs. Statistical significance compared to CNTL unless otherwise indicated. All bar plots show mean with ± SD (*p≤0.05, **p≤0.01, ***p≤0.001, ****p≤0.0001, one-way ANOVA). For western blots quantification refer to Figure 3—source data 1.