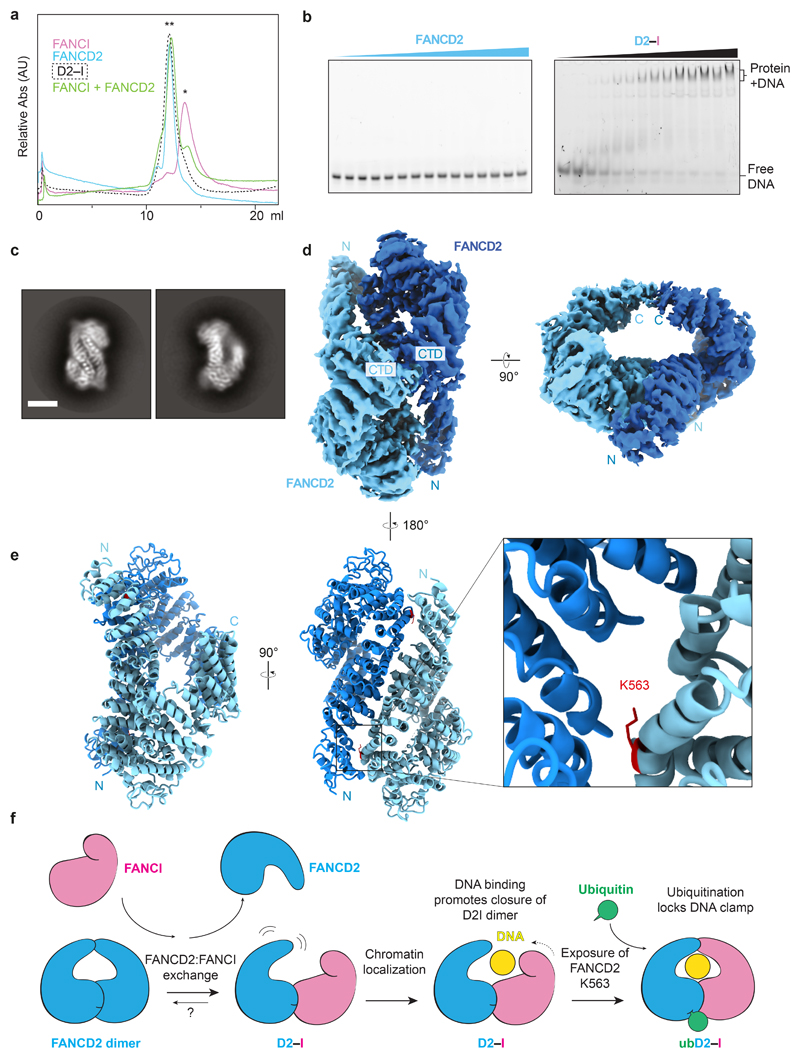
Fig. 5. FANCI forms a monomer and binds DNA whereas FANCD2 is dimeric and does not bind DNA.
a Size exclusion chromatography analysis of purified FANCI, FANCD2, D2–I, and FANCI mixed with FANCD2. A single asterisk (*) indicates the migration position for monomers. A double asterisk (**) indicates the migration position for dimers. This experiment was performed three times and a representative chromatogram is shown. b DNA binding of FANCD2 and D2–I was analyzed by EMSAs performed with 20 nM 39 bp double-stranded DNA and 0–140 nM protein. Representative gels of experiments independently performed three times are shown. Uncropped gels are available in Supplementary Fig. 1. c Selected 2D reference-free class averages of FANCD2. Scale bar is 100 Å. d CryoEM map of FANCD2 homodimer. The locations of the N- and C-termini are marked. e Model of FANCD2 dimer. The buried K563 residue (red) on FANCD2 is shown in close-up. f Model for regulation of FANCD2 and FANCI in DNA crosslink repair. Isolated FANCD2 purifies as a homodimer that is closed, does not bind DNA, and is not monoubiquitinated. Upon incubation with purified (monomeric) FANCI, this exchanges into a D2–I complex with an open conformation. D2–I binds and encircles DNA, converting the complex into a closed conformation, and thereby acting as a DNA clamp. The ubiquitination site on FANCD2 is exposed in the closed conformation, allowing access to the FA core complex and E2 enzyme. Ubiquitin locks the D2–I clamp in a closed conformation so it is not readily released from DNA.