Figure 4: Functional changes of the gut microbiome and altered arginine metabolism in the PAH cohort.
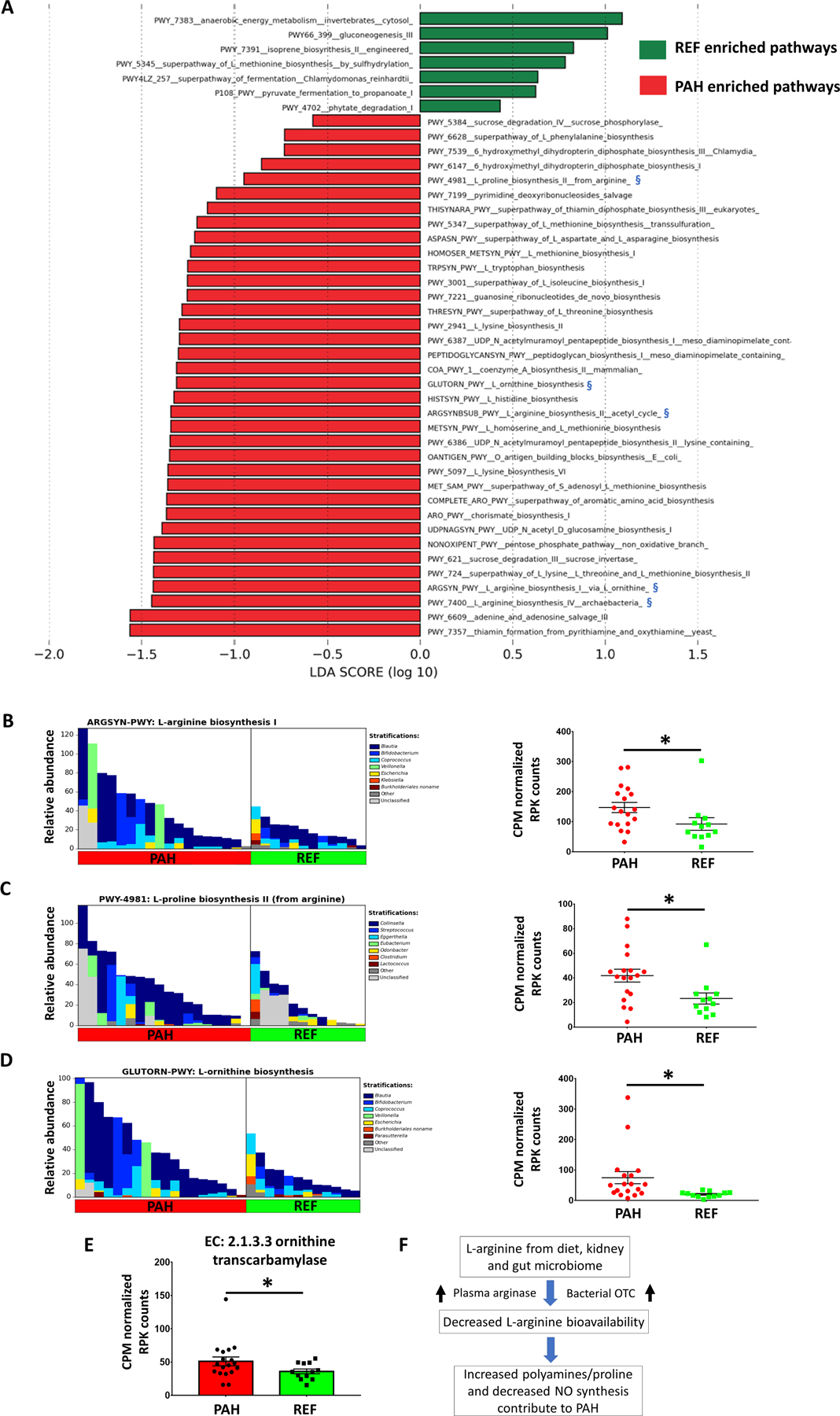
(A) LEfSe of altered functional pathways of the microbiomes in PAH and REF cohorts. Biosynthesis of several amino acids was increased in PAH such as arginine, lysine, homoserine, methionine, ornithine and tryptophan. Arginine/proline/ornithine biosynthesis was increased in PAH microbiome and marked with blue symbols (§). (B) Genera contributing to L-arginine biosynthesis (left) and CPM normalized RPK counts for the respective pathway by MetaCyc analysis (right). (C) Genera contributing to L-proline biosynthesis (left) and CPM normalized RPK counts for the respective pathway (right). (D) Genera contributing to L-ornithine biosynthesis (left) and CPM normalized RPK counts for the respective pathway (right). (E) Ornithine transcarbamylase (OTC), which converts L-ornithine to L-citrulline, was significantly increased in PAH microbiome. (F) Schematic diagram of potential bacterial contribution to PAH pathogenesis. Along with increased plasma arginase, bacterial OTC may contribute to decreased arginine bioavailability in PAH47.
