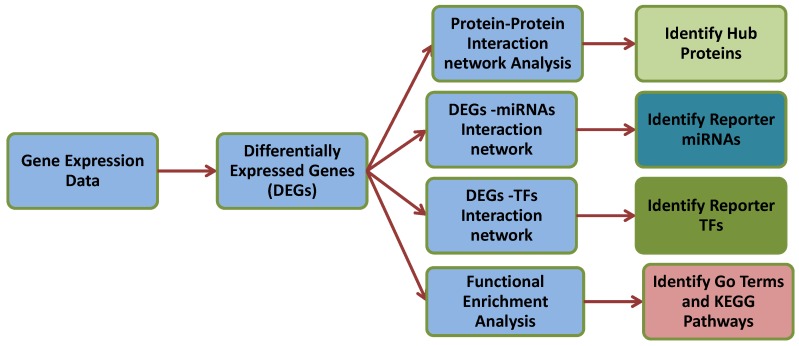
Figure 1.
The multi-stage analysis methodologies employed in this study. Gene expression datasets related to follicular thyroid carcinoma (FTC) and follicular thyroid adenoma (FTA) tissues were collected from the NCBI Gene Expression Omnibus (NCBI-GEO) database and statistically analysed using GEO2R to identify differential expression of genes (DEGs). Four types of functional enrichment analyses of DEGs were then performed to identify significantly enriched pathways. Thus, we constructed protein-protein interaction networks around DEGs topological analyses to identify putative pathways hub proteins, identified possible micro-RNA (miRNA) and transcription factor (TF) interactors, and used Gene Ontology annotation terms to provide pathways enrichment. TF and miRNA studies employed JASPAR and miRTarbase databases, respectively. DEGs were integrated with those networks, and higher degree and the betweenness centrality were used to designate TFs and miRNAs as the reporter transcriptional regulatory elements. The target DEGs of reporter miRNAs and TFs were subjected to pathway enrichment analyses.