Figure 3.
Gaf1-Dependent Gene Expression
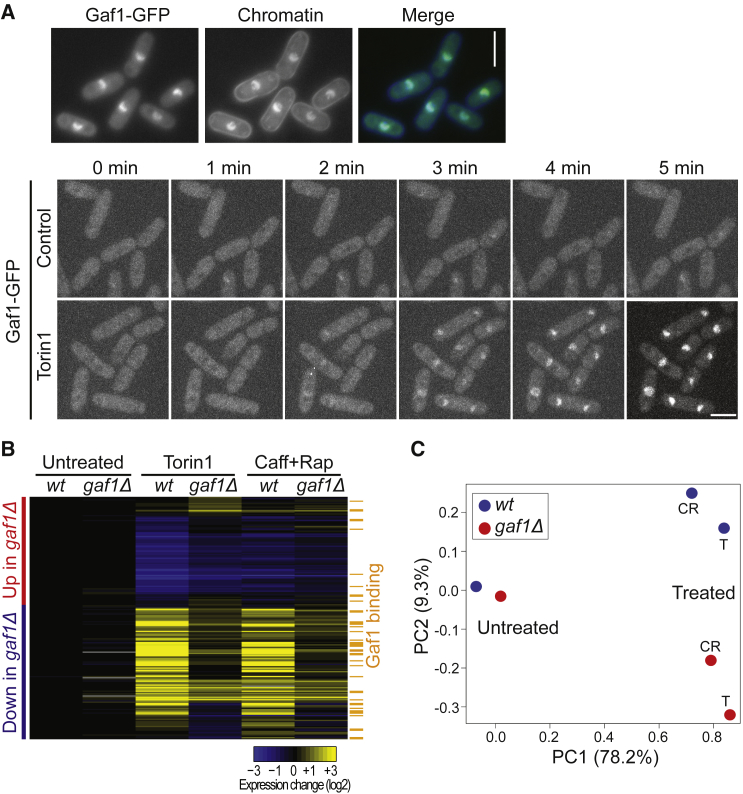
(A) Top panels: fluorescence microscopy of cells expressing GFP-tagged Gaf1 (left) with chromatin stained by Hoechst 33342 (middle) after 10 min of exposure to 20 μM Torin1. Bottom panels: fluorescence microscopy of live Gaf1-GFP cells, showing stack projections of 1-min time lapses in rich medium. Cells are shown before (0 min) and in 1-min intervals after addition of either DMSO (solvent control; upper panels) or Torin1 (20 μM final; lower panels). Gaf1-GFP is visible inside the nucleus within 3 min after Torin1 addition. Scale bars: 5 μm. See also Figure S2.
(B) Hierarchical clustering of microarray data. Columns represent WT or gaf1 mutants (gaf1Δ) before (untreated) and after 1 h of treatment with 20 μM Torin1 or with 10 mM caffeine and 100 ng/mL of rapamycin (Caff+Rap). Rows represent the 198 genes whose mRNA levels changed ≥1.5-fold in Torin1-treated gaf1Δ cells relative to WT cells, consisting of 90 genes showing higher expression (red bar) and 108 genes showing lower expression (blue bar) in gaf1Δ cells. In untreated cells, only 3 genes showed ≥1.5-fold expression changes in gaf1Δ relative to WT cells. Average RNA expression changes (from 2 independent repeats) in the different genetic and pharmacological conditions relative to WT control cells are color coded as shown. The orange bars indicate 43 genes whose promoters were bound by Gaf1 after 60 min with Torin1. See also Figure S3.
(C) Principal-component (PC) analysis of all genes measured by microarrays. PC1 separates untreated cells from cells treated with Torin1 (T) or caffeine and rapamycin (CT), while PC2 separates WT (blue) from gaf1 mutants (gaf1Δ, red). Percentages of the x and y axes show the contribution of the corresponding PC to the difference in the data.