Figure 4.
Gaf1 Regulation of Protein-Coding and tRNA Genes
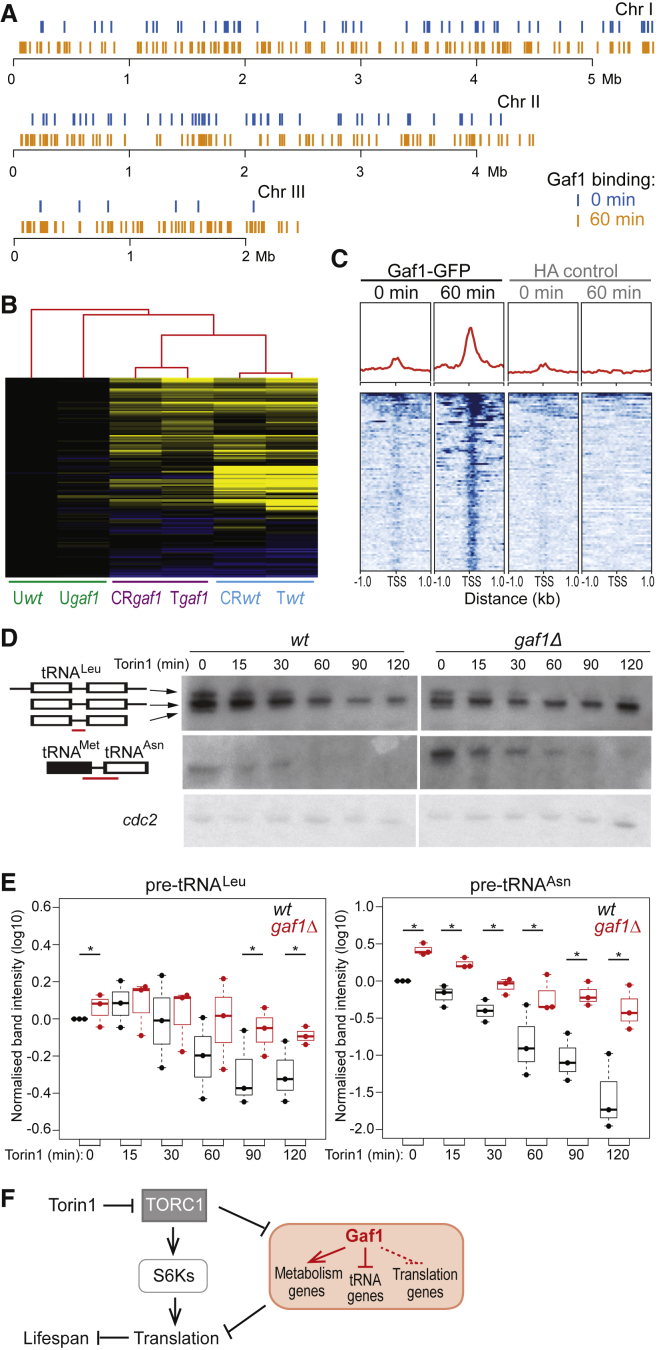
(A) Gaf1 binding sites across the 3 S. pombe chromosomes, before (0 min, blue) and after (60 min, orange) treatment with 20 μM Torin1. See also Figure S4.
(B) Hierarchical clustering of microarray data for 150 protein-coding genes bound by Gaf1 after Torin1 treatment and for which expression data were available for all conditions. The conditions have been clustered as well (red tree on top) and are grouped as follows: untreated WT and gaf1Δ cells (Uwt, Ugaf1), caffeine+rapamycin- or Torin1-treated gaf1Δ cells (CRgaf1, Tgaf1), and caffeine+rapamycin- or Torin1-treated WT cells (CRwt, Twt). Expression changes are color coded as in Figure 3B.
(C) Gaf1 shows increased binding to tRNA genes after Torin1 treatment. Top, red curves: average Gaf1 binding profiles aligned to transcription start sites (TSSs) of all S. pombe tRNA genes before (0 min) and after (60 min) Torin1 treatment, along with corresponding control ChIP-seq data (hemagglutinin [HA]). Bottom: heatmaps of Gaf1 binding around the TSS of all 196 tRNAs, ordered by normalized ChIP-seq coverage. See also Figure S5.
(D) Northern blots of precursor tRNAs for leucine (top) and asparagine (middle) from WT and gaf1Δ cells, treated with 20 μM Torin1 over 120 min as indicated. Probes to detect precursor tRNAs, indicated with red bars, are as described before (Otsubo et al., 2018). Probes for cdc2 were used as loading control (bottom).
(E) Northern quantitation of leucine and asparagine precursor tRNAs relative to WT time 0 and normalized to loading controls (three independent repeats shown as dots). Asterisks denote significant differences in pre-tRNA levels between WT and gaf1Δ cells from same time point (t test, p < 0.05).
(F) Model for Gaf1-mediated transcriptional repression of translation downstream of TORC1. Following TORC1 inhibition, Gaf1 activates the transcription of genes for small-molecule metabolic pathways and represses the transcription of tRNAs and other genes functioning in translation (the latter via indirect control, hatched). Together with the S6K-mediated translational control (Ma and Blenis, 2009), this transcriptional branch contributes to longevity.