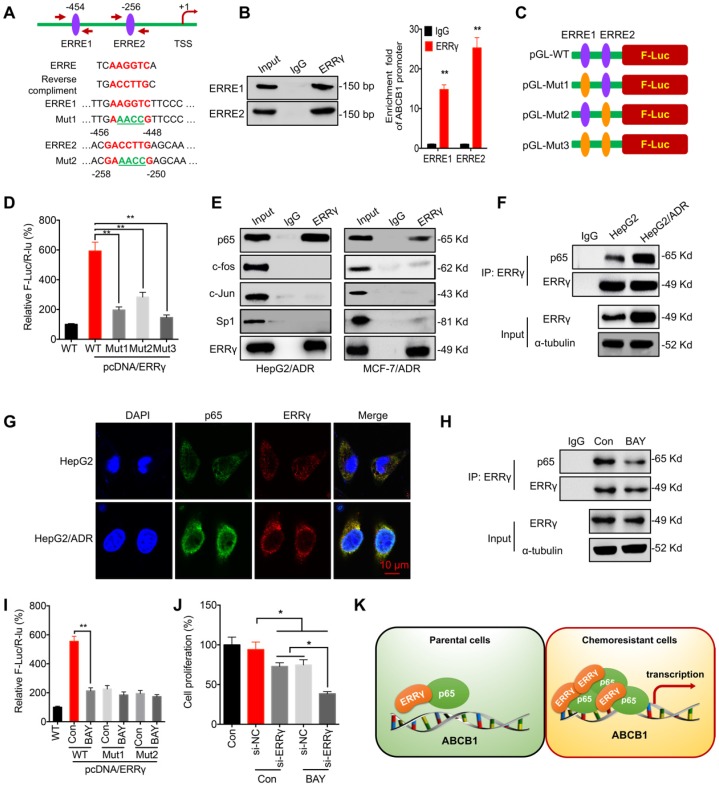
Figure 4.
ERRγ interacts with p65 to regulate ABCB1 transcription. (A) Schematic representation of ERREs in the promoter region of ABCB1 with changes of nucleotides in ERRE1 and ERRE2 shown as indicated; (B) ChIP-PCR assay showing ERRγ binding to ERRE1 and ERRE2 in ABCB1 promoter. The input (5%), binding between ERRγ and the promoter of ABCB1 at the potential binding site ERRE1/2, was amplified by qPCR (right) and confirmed by 2% agarose gel electrophoresis (left); (C) Schematic representation of mutated ERRE positions in pGL-ABCB1 vector; (D) Reporter gene assay performed in HepG2 cells 24 h post transfection with pGL-ABCB1-WT or pGL-ABCB1-Mut1/2/3 by dual-luciferase analysis; (E) Examination of ERRγ interaction with different transcription factors in HepG2/ADR and MCF-7/ADR cells following immunoprecipitation with ERRγ or control antibody and analyzed by Western blot analysis; (F) Interaction between ERRγ and p65 in HepG2 and HepG2/ADR cells monitored by immunoprecipitation using anti-ERRγ antibody; After ERRγ was immunoprecipitated, the binding between ERRγ and p65 was examined by Western blot analysis. An equal amount of ERRγ was loaded for normalization according to a pre-Western blot; (G) Expression and localization of p65 (green) and ERRγ (red) in HepG2 and HepG2/ADR cells visualized by confocal imaging; (H) Interaction between ERRγ and p65 in HepG2/ADR cells treated with or without BAY 11-7082 for 12 h and then analyzed by immunoprecipitation using an antibody against ERRγ; (I) Dual-luciferase reporter gene assay performed in HepG2 cells transfected with pGL-ABCB1-WT or pGL-ABCB1-Mut1/2, with or without pcDNA/ERRγ, for 12 h and then further treated with or without BAY 11-7082 for 12 h; (J) HepG2/ADR cells were treated with si-RNA or si-ERRγ combined with or without BAY 11-7082 for 12 h and then further treated with 5 μM Dox for 48 h; (K) Model for ERRγ/p65-promoted transcription of ABCB1 in chemoresistant cancer cells. Data were presented as means ± SD from three independent experiments. **p< 0.01. NS, no significant.