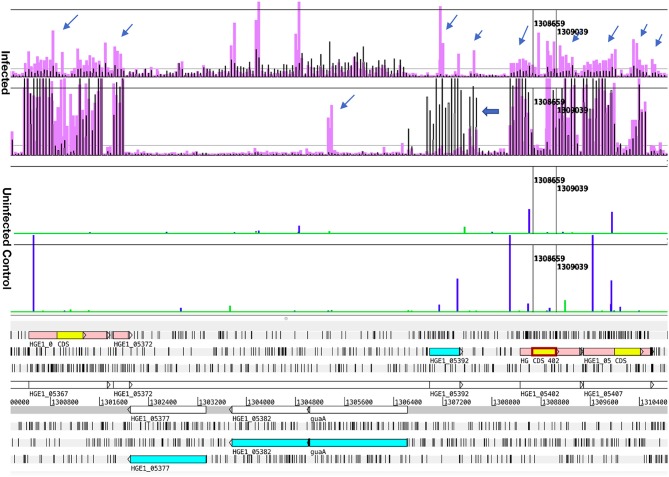
Figure 3.
Comparison of transcript signal from Samples 1 and 3, illustrating antisense transcripts and transcription from an unannotated gene. Transcription data (top four graphs) plotted over the annotated genome sequence (sections at the bottom; turquoise boxes denote coding regions of annotated genes, pink boxes denote p44 conserved sequences with yellow boxes denoting their center hypervariable region) of A. phagocytophilum (Ap) strain HGE1 using the Artemis genome browser. The top two graphs (Infected) represent Ap transcription in Sample 1 (black bars, 2 h with HL-60 cells) vs. in Sample 3 (lavender bars, 2 h with ISE6 cells). Each vertical bar corresponds to one 60-mer probe on the tiling array. The two graphs below (Uninfected control) are plots of data from uninfected cells (HL-60, green; ISE6, blue), to demonstrate that host cell transcripts are not responsible for antisense signals. Occasional probes produce signals but overall the sterile cell graphs are flat. Prominent antisense signals generated by Ap from ISE6 are indicated by slender arrows, including those associated with the p44 conserved regions (pink boxes). Also shown is a p44 paralog (HGE_05367; at base position 1,300,800 in the genome) specifically transcribed in Sample 3, as indicated by signal corresponding to the center hypervariable region (yellow box). Note a hypothetical gene, HGE1_05392, with elevated transcript levels in Sample 1 (and with antisense signals in Sample 3). Immediately downstream, a transcription peak indicates the presence of a small, unannotated gene (bold arrow). Transcription levels from the HVRs of p44s HGE_05367, HGE1_05402, and 05407 were rated 1, 0, and 2 in HL-60, and 4, 0, and 1 in ISE6, respectively. Nucleotide positions in the genome are indicated by numbers above the gray line.