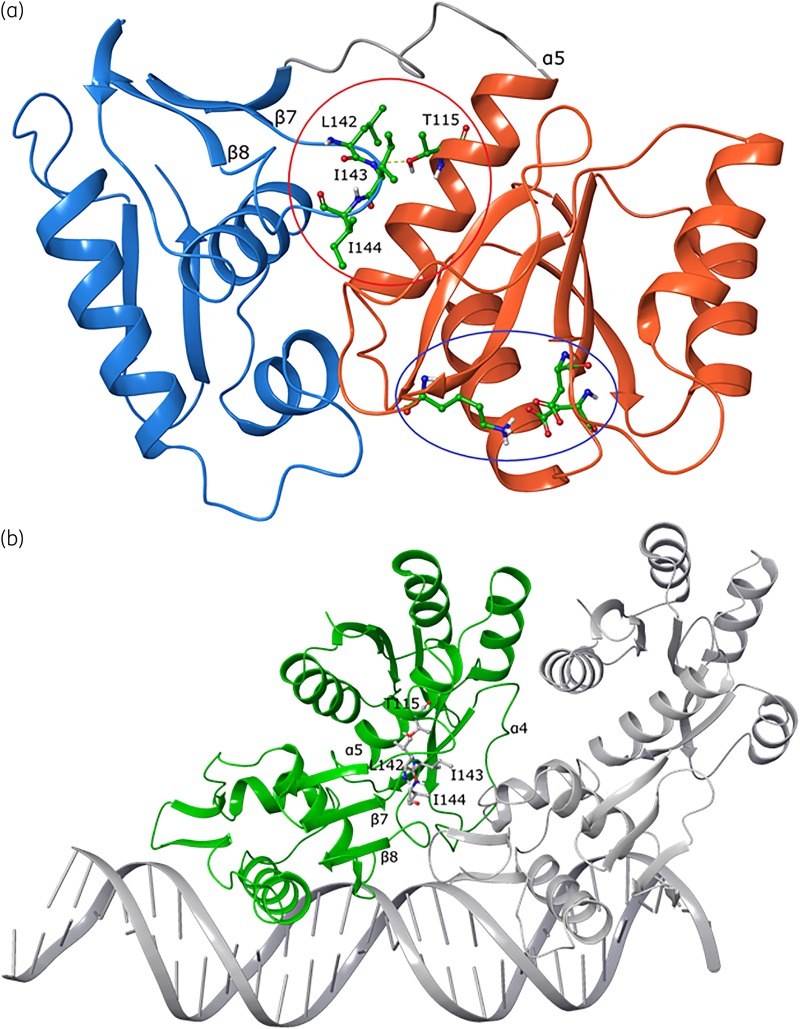
Figure 5.
Structural analysis of mutant VanRCd in monomeric and DNA binding conformations built from the M. tuberculosis PhoP structure. (a) Monomeric VanRCd shows the DNA binding effector domain in blue ribbons, with the key helix that engages DNA at the far left. The receiver domain is shown in orange ribbons, with the conserved phosphorylation site (lysine and aspartic acid residues) shown as balls and sticks with green carbons (indicated by the blue oval). The resistance-conferring Thr115 substitution site is on the receiver domain and its interaction with a key loop containing lipophilic residues (Leu142, Ile143 and Ile144) on the effector domain is shown; the loop is indicated by the red circle and the residues are shown as balls and sticks with green carbons. (b) Interaction of VanRCd (monomer A, green) and second monomer B (PhoP, grey) with a segment of DNA. Shown as ball and stick representations are the Thr115 amino acid site on the receiver domain and interacting residues from the adjacent loop on the effector domain.