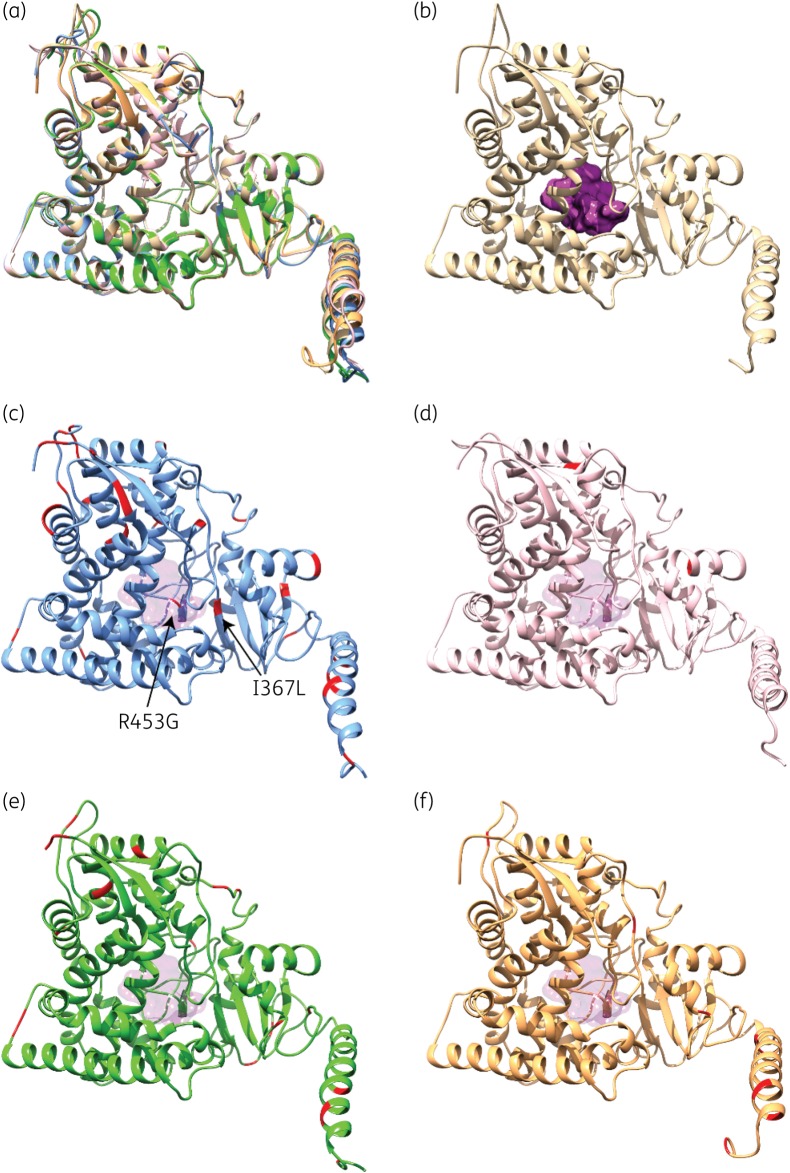
Figure 3.
Structural homology models of the five CYP51A variants investigated in this study. (a) Structural overlay of CYP51A models from HMR-AF-270 (beige; azole-resistant A. fumigatus with WT CYP51A), HMR-AF-23 (blue; azole-susceptible A. turcosus), HMR-AF-1038 (pink; azole-resistant A. turcosus), HMR-AF-39 (green; azole-resistant A. thermomutatus) and HMR-AF-1185 (orange; azole-resistant A. lentulus). (b) 3D homology model of WT CYP51A from HMR-AF-270, highlighting the buried active-site cavity (purple surface). (c) 3D homology model of CYP51A from HMR-AF-23, highlighting silent amino acid replacements diverging from the WT A. fumigatus sequence (in red). (d–f) 3D homology models of CYP51A from HMR-AF-1038 (d, pink), HMR-AF-39 (e, green) and HMR-AF-1185 (f, orange), highlighting amino acid replacements diverging from the WT A. fumigatus sequence (in red). For positional comparison, active-site cavity surface transparency is shown in all CYP51A variants.