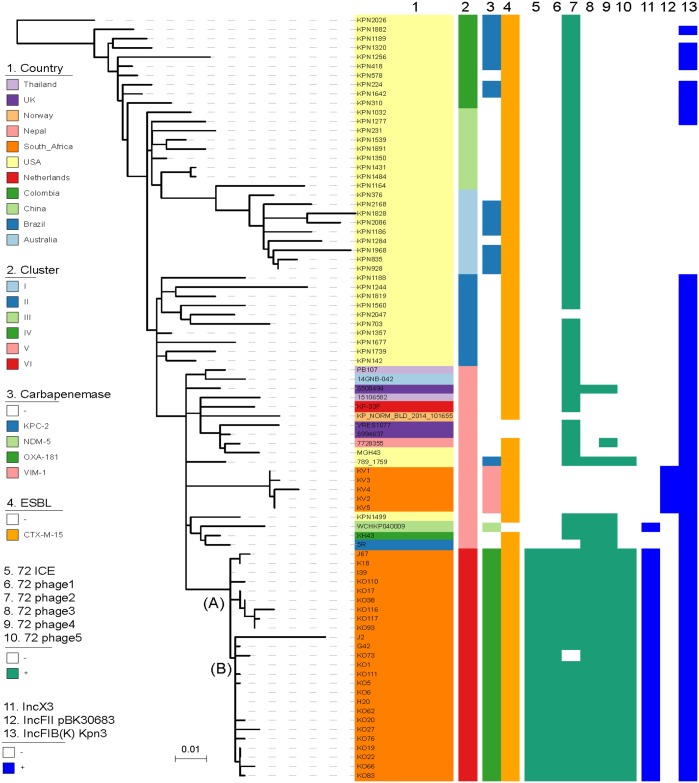
Figure 2.
Phylogenetic analysis of K. pneumoniae ST307 from TTH including some representative global ST307 isolates to highlight different clades. Isolates from TTH (n = 24), isolates from private Pretoria hospitals (n = 6) and global isolates (n = 53) are included. The global genomes were obtained from the NCBI WGS database. Clades and lineages were defined using previously published criteria.4 OXA-181 ST307 isolates belonged to a distinct South African clade, namely clade VI. Analysis identified two distinct lineages among this clade, namely ST307_181A (A) and ST307_181B (B), as illustrated at the nodes. Isolates J67, K18, I39, J2, G42 and H20 were obtained from a previous study involving private South African hospitals.4 Isolates KO110, KO17, KO38, KO116, KO117, KO93, KO73, KO1, KO111, KO5, KO6, KO62, KO20, KO27, KO76, KO19, KO22, KO66 and KO83 were from this study. VIMST307 (KV1–KV5 from this study) belonged to a different ST307 clade (cluster V) that clustered with other international ST307. ST307 clades I–IV are limited to the USA. This figure appears in colour in the online version of JAC and in black and white in the print version of JAC.