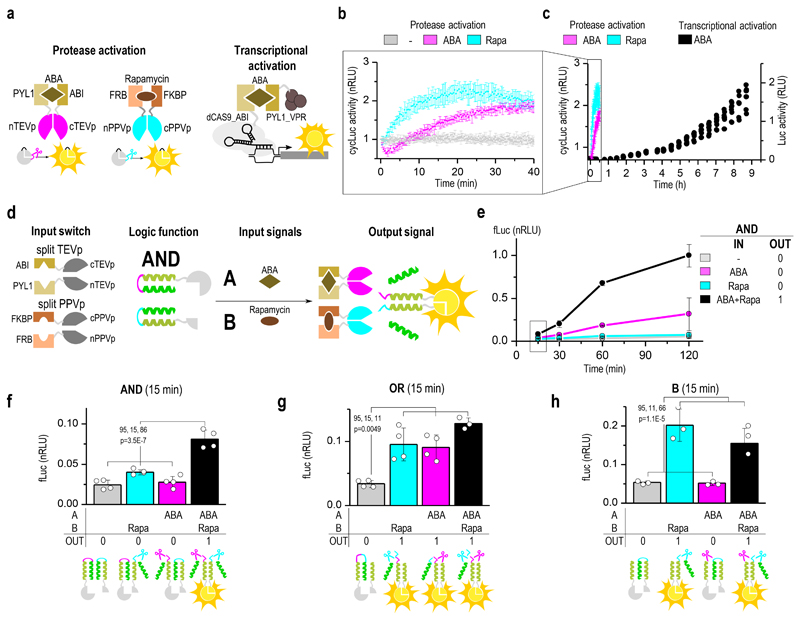
Figure 5. Fast kinetics of the proteolysis-mediated signaling pathway.
(a) Scheme of the reconstitution of the split tobacco etch virus protease (TEVp) by abscisic acid (ABA) and split plum pox virus protease (PPVp) by rapamycin measured using cyclic luciferase reporter in HEK293 cells. A CRISPR/dCas9-based activator was used as comparison for the kinetics of the transcriptional control of luciferase activation. (b) Continuous online monitoring of chemically induced luciferase activity in HEK293T cells and comparison with the kinetics of transcriptional activation (c). (d) Schematic presentation of building blocks for inducible split protease-cleavable orthogonal CC-based logic (SPOC logic) functions. (e) Kinetics of chemically regulated AND function where both segments of the split luciferase are activated by proteolytic cleavage. Significant response was detected in less than 15 minutes after the addition of chemical inducers. (f-h) SPOC logic functions AND, OR, and B regulated by rapamycin and ABA 15 minutes after induction. Transfection mixtures are listed in Supplementary Table 5, 6. Values are the mean of three (f) and four (b, c, e-g) cell cultures ± (s.d.) and are representative of two independent experiments, significance tested by 1-way ANOVA with Tukey’s comparison between the indicated ON and OFF states (values CI, df, F and p are indicated on graphs (f-h)).